A new review article on Evolutionary causes and consequences of gene duplication has dropped. It’s nothing novel to well-informed biologists, but it’s another nail in the coffin of creationism. Not that they will care; we’ve been explaining that common genetic mechanisms can routinely increase the information content of the genome, and that we can witness how new genes with new functions arise, and it never sinks in.
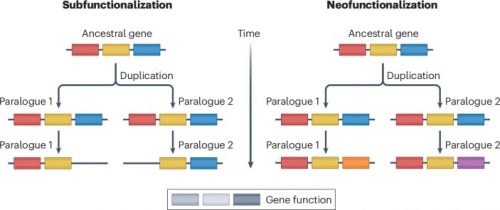
Gene duplication is the primary mechanism by which new genes emerge. Models and empirical studies have shown that paralogous genes are maintained because of dosage benefits, the partitioning of ancestral functions or the acquisition of new functions. However, the underlying molecular mechanisms and the relative importance of the factors driving evolution towards one fate or another have remained difficult to quantify. Recent advances in experimental and computational methods, such as gene editing, deep mutational scanning and ancestral sequence reconstruction, have enabled molecular analyses of duplicated gene evolution across timescales. Combined, these approaches are revealing how adaptive and non-adaptive evolutionary forces shape the modern fates of gene duplicates.
I imagine some might leap on the phrase “remained difficult to quantify,” but that’s the point of the paper: new techniques have been developed that allow us to quantify those details. The review specifically brings up multiple examples.
Divergence in interaction specificity following duplication has profound consequences on cell biology. For instance, the neofunctionalization of steroid receptors, a family of hormone-activated transcription factors with roles in development and stress responses, evolved following multiple rounds of WGD[whole genome duplication] in vertebrates. Although one paralogue maintained its ancestral interactions, the other acquired mutations, conferring on it the capacity to bind different hormones and DNA motifs. Studies of transcription factors in plants, yeast and other organisms have identified many paralogues that diverged in their specificity for transcription factor binding sites and distal regulatory elements. Such divergence in interaction specificity has enabled multiple species to acquire novel regulatory modules over time.
The conclusion discusses some of those mechanisms.
Evolutionary biologists have long been interested in the fate of duplicated genes. Long-standing questions include which factors promote the fixation or long-term retention of duplicates, and their divergence in terms of sequence, expression, interactions and function. Multiple emerging technologies have enabled directly testing how adaptive and non-adaptive forces drive the evolution of paralogues. For example, fitness functions derived by tuning expression level with synthetic biology tools have enabled testing whether increases in protein abundance due to duplications are beneficial or not. Deep mutational scanning and comparisons between extant and reconstructed pre-duplication ancestral sequences facilitate the identification of mutations that alter a particular function. In particular, comparisons between different paralogues have shown that the fixation of function-altering mutations is often contingent on the presence of other mutations that originally had no effect on fitness. Similarly, other paralogues can become dependent on each other if their heteromers become the only functional unit. Therefore, multiple sources of evidence highlight the role of non-adaptive processes in the evolution of duplicated genes.
Continuing to combine and develop new methodologies will help to address open questions about the fates of paralogues. Although the likelihood of functional divergence between paralogues increases with the age of the duplication, the time required to reach functional divergence might vary depending on the pair of paralogues. In fact, multiple underlying factors may contribute to variation in the rate of functional divergence, such as the type of function performed by the paralogues. In turn, progressive changes in functions such as catalysis and binding specificity are likely to modify the fitness functions of the paralogues, allowing natural selection to distinguish between them. Ultimately, assaying such subtle and progressive mutational effects on gene function will help to better trace the evolutionary history of paralogues and the forces that shaped them.
It’s a nice summary of the problems and potentials for studying evolutionary gene duplications. I’m adding it to my list of papers to study in greater depth.
Creationists will pretend it doesn’t exist.
Angel F. Cisneros, Soham Dibyachintan, Frédéric Bédard, Simon Aubé, Pascale Lemieux & Christian R. Landry (2026) Evolutionary causes and consequences of gene duplication. Nature Reviews Genetics https://doi.org/10.1038/s41576-026-00935-5.


How much a tendency for gene duplicates to decay into nonsense and add to the junk in our trunk?
One thing that fascinated me is that elephants apparently have more copies of p53, helping them to keep cancer in check.
But you cannot randomly insert genes without risk of adverse effects so the ancestors of elephants have not only duplicated those genes, they must have dealt with copies situated in bad locations.
Unfortunately my memory is not good enough to benefit from a course in modern genetics. There is too much to take in.
Hemidactylus #1
Can’t speak in terms of numbers, but the fact that you now have two copies of a given gene will mean that one of said copies can be broken with no deleterious effect on the organism. Thus, more potential for junk.
LykeX #3
Maybe, but the article mentions one case in which multiple identical copies can all be subject to selection: dosage benefits.
In one sense, genetic information change is easy to quantify. You can calculate the exact Shannon complexity of any given sequence of symbols, including gene or protein sequences. What the authors of this article are getting at, of course, is that it is difficult to quantify the developmental and evolutionary impact of these changes, largely because genetics within a population of organisms is huge, complicated, and influenced by the environment.
Of course creationists don’t want to understand any of this. They’d rather repeat long-debunked statements like “mutations can’t increase complexity.” Basically, they make up “facts” about genetics, evolution, and information theory while refusing to educate themselves to a barely introductory level in any of those fields.
BTW, that link is only for UMN staff and students. The paper can be found here in Nature Review Genetics. Unfortunately, everything but the abstract is behind a paywall for anyone without academic access.
The genetic building blocks underlying all life on Earth are simply mind boggling.
That their wonder has (only) been revealed by patient, careful, persistent reiterative application of the Scientific Method through experimentation, and not pure philosophy (including, generously, theology) says. it all, doesn’t it?
Many people’s reverence is wildly misplaced.