Update
I’m much better in terms of hypervigilance. But it’s still useful to keep my number of social spaces to a minimum while I think about social variables around me. “Social firmware update”? It makes sense that groups would complicate that. “Gender-Null” still makes sense. It’s easy to just be “like everyone” when the group is more sensitive to the words than me. I make choices with the words. Relative to tourette syndrome maybe it’s an “intense words” thing. I make choices with lots of words that other people feel intensely about. This is not to say that tourette syndrome and non-binary or gender-null are connected, rather these parts might have connections for me.
I can’t say this is me going back to blogging on a regular basis either. This is more like I have a creative project connected to a lot of feelings about what I wish I was doing professionally, origin of life research. Obsessing over brain science is more of a defence mechanism, this is an older obsession and it’s not only interesting but I may get some benefits out of it. Lots of feelings here.
I have spent a lot of time drawing metabolic pathways until I can do it from memory as a way of internalizing them. More body parts and sensations involved enhances memory. Drawing anatomy is helpful too. And I’ve had a lot of time to think about the patterns. I’ve come back to one project over and over and I recently got a tablet with a good screen for drawing and transferred the latest version of a group of pathways to Sketchbook. I’ve also started using Kingdraw for molecular structures.
BUT, I can’t just post this drawing by itself. If you have a strong background in molecular biology you can skip to the end (check out the transcription and translation diagram though, I integrated aaRS class information in with the genetic code) but for everyone else I need some figures to make this somewhat more meaningful. An attempt to go from public perception to hardcore molecular biology.
This is for fun. I’m playing with the ideas and concepts. So there is a lot of me imagining things in here but I try to be clear about what is data and what is me speculating. Other views are welcome, including data that suggests I’m wrong, or people who have made the same observations that I missed. This is a work in progress, but keep in mind that this is about fun.
From public perception to molecular biology.
The perception about A G U C T nucleic acids are usually understood by their relationship with amino acids through the genetic code and the making of “proteins”. Equating the metaphorical “hard drive” with its products hides the full range of relationships that must be taken into account, even pinning things relative to DNA sequence or protein sequence.
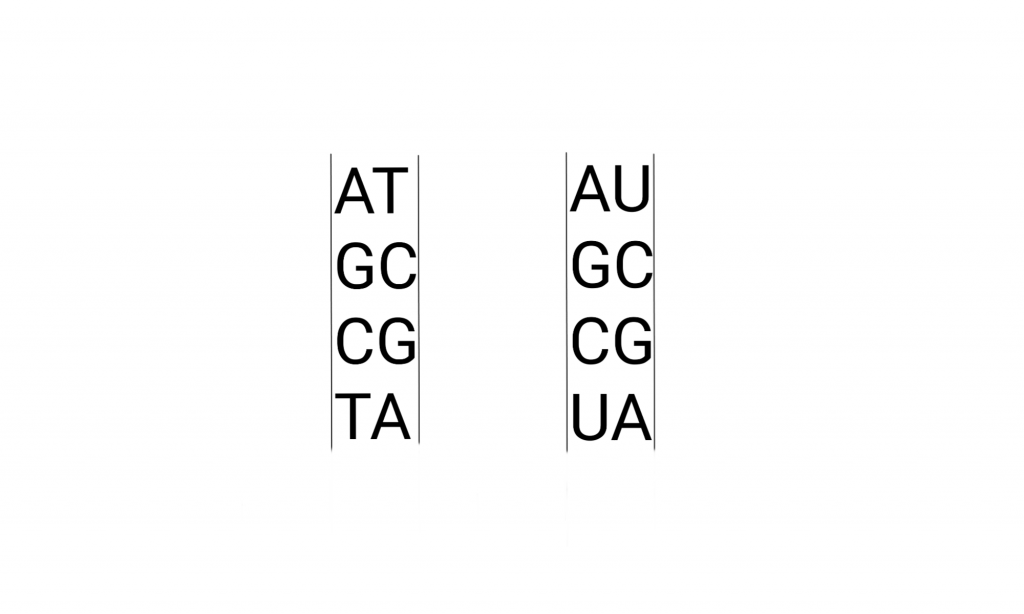
The letters pop up all over culture, but usually not the “U”. Here’s what that looks like in detail, the drawing includes the pathways that make these molecules. And metabolically related molecules.

Briefly, the Deoxy-Ribo-Nucleic acids are “duplex chains” of -ribose-phosphate-ribose-phosphate- with Adenine, Guanine (purines), Cytosine, or Thymidine (pyrimidines) attached to the Ribose by the 1′ Carbon (read as “one prime carbon” ). The structure of the “nitrogenous bases” is such that attractions (hydrogen bonds) between molecules allow A:T and G:C pairing between the strands.
Triplets in those chains (with 3 reading frames on each strand) are a basis of the “genetic code” used to synthesize the proteins in a cell. A “protein” is a chain of amino acids.
Below is the simplest amino acid, Glycine, and water in even more detail so I can show some more molecular features that can be referenced in the future. The previous figure omitted them and following ones will as well to simplify things.

The previous base-pairing diagram omitted parts of molecules worth mentioning that aren’t often represented.
- Ionic charges. Many of these molecules are actually ions in water, amino acids lose a proton (hydrogen nuclei) on one end, and gain one on the other (center versus right). This comes down to some atoms having more “electronegativity” than others (nuclei attract electrons more strongly) Most of the time I don’t do this to save effort.
- “lone pairs”. Oxygen and nitrogen atoms have electrons in “non-bonding orbitals” (the places electrons go). These regions are more negative while not being full ionic charges, called “polar” . That’s why nitrogen atoms can either attract or repel one another in the base pairing. If a hydrogen is sticking out from one base and a lone pair from another they attract.
- Water also has lone pairs, it’s actually a pyramid shaped, highly polar molecule. There is no “hydrophobic”, water is attracted to itself and crowds non-polar molecules away as it is attracted to itself. I also mostly omit lone pairs to save effort.
Now for all of the amino acids your body, and all cells, use to make proteins.

Cells use these 20 amino acids to chain together “poly-peptides” (the bonds are called peptide bonds) that do biochemistry or make structures. Essentially they are glycine + something at a specific carbon. (There’s a layer called chirality I’m not getting into yet, the hydrogens on that carbon aren’t the same chemically and biology uses one here).
This is my own take on how to organize them. There are 2 organizing principles; metabolic relationships, and atoms in the side chain.
The untitled block on the left roughly acts as precursors for ones to the right. There’s a system that can generate serine from glycine and back as the cell needs. Serine itself can be used to make cysteine. Larger amino acids can be broken down into smaller ones too.
The “aromatic amino acids” (Phe, Tyr, Trp, named for the benzine ring) are complicated because they are made from 3 carbon and 4 carbon molecules. So I put them on a different level.
Tyrosine and histidine are close because they both use ribose in their biosynthesis.
Making proteins from DNA.

This is a simple protein made with the previous “proteogenic amino acids” (cells use these). The next figure will explain how, and why methionine is first.
This is how cells make proteins with DNA sequences.
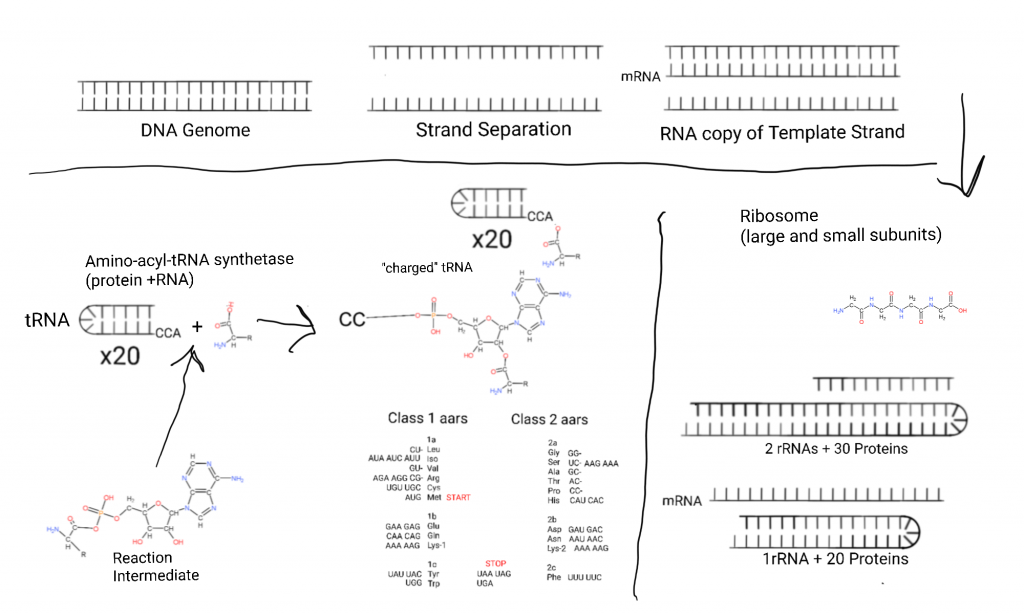
This figure starts on the top left with the DNA genome, so named because it contains the “genes” expressed into proteins (now more complicated due to RNA genes). Some cells have “linear” chromosomes of 100’s of millions of base pairs (genome segments), and bacteria tend to have small genomes of a few million.
The DNA is separated into it’s 2 strands and one strand acts as a template for copying a part or all of the genome. That copy of a part is made from RNA in a process called Transcription. The copy is called an “mRNA” or “messenger RNA”. Many proteins are involved.
The mRNA is escorted to the smaller half of a massive 2 subunit complex called the Ribosome where the nucleic acid sequence is transformed into a poly-peptide sequence. The small subunit (SSU) contains an RNA called rRNA or “ribosomal RNA” and 20+ proteins in bacteria.
After the mRNA docks with the SSU a large subunit (LSU) made from 2 rRNAs and 30+ proteins in bacteria docks with the small subunit.
Small (50-70 nucleotides, nts) RNAs called transfer RNAs (tRNA) bring in amino acids one at a time and a 3 nucleotide sequence in the mRNA called a “codon” determines what amino acid is placed through binding to an “anticodon” on the tRNA. (In the bottom-left I show an intermediate in the aminoacylation of tRNA, an aminoacyl-AMP intermediate.)
Both protein and RNA are involved in aminoacylation.
A 3 codon system gives 64 possibilities and the way those are arranged is the generic code. 3 of these are reserved for STOP: UAA, UAG, and UGA. Methionine doubles as START with AUG.
There are 2 classes of tRNA and systems of attachment of amino acids to tRNA. The aminoacyltRNA transferase systems. Class 1 involves attachment of amino acids to the 2′ hydroxal of ribose, class 2 involves attachment of amino acids to the 3′ hydroxal of ribose. On the figure I have listed the classes and subclasses under the relevant hydroxals and listed their codons next to them.
The ribosome subunits hold the mRNA, and the tRNAs in place and one amino acid is added at a time while empty tRNAs leave. Eventually a STOP is reached and a new poly-peptide/protein is released from the exit tunnel of the large subunit.
These proteins are then often modified in many ways and transported to many places to do their structural or catalytic jobs.
The genetic code relationship plus ribose metabolism.
That is not ONLY how I see these things. I also see the individual biochemical reactions that cells use to make nucleic acids. Starting with RNA. (Anabolism=construction, catabolism=breaking down and I will need to look at catabolism at some point).
DNA as a physical substrate for molecular biology, our “hard drive” is based in RNA metabolism. And so the metabolism of RNA, not deoxy-ribose is context to understand these relationships. (Deoxy-nucleotides are definitely part of the story though.)
Ribose centered metabolism.

Note that this is at lower resolution but still ok for display. A full resolution version can be found HERE.
This is a Ribose centered metabolic diagram. A tool for thinking about how metabolism can tell us where nucleic acids and proteins came from. They have metabolic relationships beyond the DNA>RNA:>Protein one. These are parts of that relationship.
All of the hydrogens are implied to save space and work. Each protein is named with type instead of written so they stand out. Each protein is positioned with the molecules they interact with, and are products of the previous protein(s).
This is a prokaryote centered metabolic diagram. It’s been difficult to get interested in us bacterial-archal fusions (eukaryotes) here. The blending complicates the picture.
Beneath each protein is the length in amino acids, usually from E.coli strain K12 unless E.coli was the exception relative to cells in general. I’ll get around to citing everything eventually but I can get specific things if someone wants something. I’ll put some sources of information at the end, and here and there in the text.
Along with the length I list each known subdomain within the protein and the protein domain families and superfamilies to which the domains belong. These parts of proteins are related to one another and their relationships are part of the puzzle. I go over an example with PurF in the resources section. These bits of related protein often have themes to what they do, and can potentially lead to primordial subdomains.
Black arrows point in the direction of biosynthesis.
This is a very unfinished diagram. Once I did The pathways that produce the bases in nucleic acids I kept adding Ribose related pathways and I’m not done. This is maybe the 20th iteration and I keep adding elements and reorganizing.
My intention is to upload and comment on new versions, as well as adding new categories of information as big as protein subdomains (below). There are likely errors here (PurF is a GATase class 2!!! I keep putting class 1, I used PutF as an example in the references with correct info and this will be my first correction,) and after and once this is up I’ll do another error sweep before doing more. There is fading and smudging from copying and pasting. And I’m still making decisions on how I’m going to represent things so there may be inconsistent bits.
The following pathways use ribose in the form of Phospho-Ribosyl-PyroPhosphate, PRPP.
- The Purines: produced as Adenosine-MonoPhosphate (AMP), and Guanosine-MonoPhosphate (GMP). (Starts center left with PurA, runs right and down, and forks left into AMP and GMP)
- The Pyrimidines: produced as Uridine-MonoPhosphate (UMP), Cyotosine-MonoPhosphate (CMP), and deoxy-Thymidine-MonoPhosphate (dTMP) (Starts at the bottom-left, continues to the middle, and forks into CMP and dTMP after UMP)
- Amino acids: Tyrosine (starts top-left, goes down, turns right to the center, forks right into Trp, and forks up into Phe and Tyr), and Histidine (starts top-left, moves right)
- Cofactors: Thiamine (Vitamin B1, starts upper right under histidine, pathway moves right), Nicotinamide-Adenine-Dinucleotide-Phosphate (niacin, starts lower-middle-left and moves right), Cobalamin (Vitamin B12, starts to the right of NADP, this only covers the part of B12 derived from AIR),and Tetra-Hydro-Folate (to be added).
If someone is wondering why I haven’t added the cofactor that donates the formate for Purine biosynthesis to the diagram, that is an example of going too far with treating things like dispensers of things available in earlier environments. Some organisms use formyl-phosphate. But it still needs adding. Ultimately I’m betting THF, and things like glutamine (the NH3 donor, mostly) are providing things that were environmentally available. THF is a remodeled GMP (like how histidine is a remodeled AMP?) A GMP with an intermediate from the Phe/Tyr/Trp pathway (PABA) added.
Purines and Pyrimidines
The Purine pathway produces A and G. The Pyrimidine pathway produces U, C, and T. These 2 pathways interact with Ribose in opposite fashion. A and G are built onto Ribose bit by bit out of small things and rings are closed. Pyrimidines are made by building a ring and putting them on Ribose. PRPP is also used with whole nitrogenous bases that are already finished.
It might be useful to look at the pathway like this.
Purines: PRPP + NH3 + Glycine + CH2O + NH3 + CHO3 + Aspartate – Fumerate + CH2O = IMP (pathway fork).
That aspartate-fumerate is effectively a 2-step, or indirect amination (ammonia addition) that releases a TCA cycle intermediate.
IMP + Aspartate – Fumerate = AMP
And there that weird amination again. It happens in arginine production too.
I have ideas of different places in time where aspartate and AMP accumulation occured. Maybe glycine accumulation earlier too. How to define the “start” of things?
IMP + H2O + NH3 = GMP.
Pyrimidines: CHO3 + Aspartate – H2 + PRPP – CO2 = UMP
Maybe there was aspartate accumulation with more than one use. A transition from a 5 to a 6 ring polymer (Purine intermediate AIR) leading to Pyrimidines and other things? A step is shared with arginine biosynthesis (CHO3, bicarbonate use through an intermediate called carbamoyl-phosphate).
UMP + NH3 = CMP
UMP + PO4 – O + PO4 – 2 PO4 + CH4 = dTMP
Thymidine stands apart from the other nucleotides in that it is made from Uridine-DiPhosphate (UDP) that is dehydroxylated before being methylated. Since I’m a bit RNA focused I haven’t pursued Thymidine much.
Amino Acids: Histidine and Tryptophan
In histidine biosynthesis a ribose from PRPP is connected to a nitrogen on ATP. The same nitrogen provided by aspartate. Then the ribose leaves with a C-N, and the rest of the ATP is a late intermediate of Purine biosynthesis. Adding an NH3 and some processing gives histidine.
A molecule that is a 4-carbon clone of Ribose-5-phosphate, Erythrose-4-phosphate, is combined with a 3-carbon glycolysis intermediate, Phospho-Enol-Pyrivate, to make a 7-carbon molecule that is processed into a ring before being attached to ribose via an added nitrogen. After that the resulting molecule loses carbons 1-3 of ribose as the glycolysis intermediate Glyceraldehyde-3-Phosphate, a molecule like a 3-carbon Erythrose-3-phosphate (or ribose-5-phosphate) (histidine takes the ones ribose loses here…). That G3P is replaced with a serine to make tryptophan.
Cofactors: Thiamine, NAD, and THF
A cofactor is like an “attached functional module” for proteins. Like different heads on a power tool.
There’s Thiamin (VB1), a carbon group extractor/dispenser. Interestingly AIR is made into the part of Thiamine that isn’t grabbing carbon chains. But it’s rearranged into HMP in an interesting set of reactions where the ribose is dismantled and parts are attached to the carbon-nitrogen ring to make a 6-membered ring. And the thiazole-P that HMPP is combined with looks a lot like the ring on AIR but with sulfur instead of nitrogen.
Thiamine is part of many complexes and it acts to move carbon chains from one molecule to another. In the Pentose-Phosphate-Pathway Thiamine is one way ribose-5-P is made by moving 2-carbon units between other carbohydrates (carbon with water, hydrocarbons are carbon with hydrogen). Maybe we’re descended from the thiazole?
And while it’s fuzzy on the diagram ThiO/ThiH are two routes to make what looks like glycine, but there’s a double bond on the nitrogen (an imine as opposed to an amine). Like the double bond on FGAM, the molecule right before AIR (and see NADP). There’s some interesting things with glycine similar molecules here and there.
Then there’s NAD(P) (the P is a phosphate that changes Nicotine-Adenine-Dinucleotide’s metabolic role, the 2′ hydroxal, interesting…), a proton extractor/dispenser. It’s effectively the same for electrons. Aspartate is involved again and now it gets an imine before getting combined with DiHidroxyAcetonePhosphate, another glycolysis intermediate before ring synthesis and attachment to ribose.
Then there’s cobalamin (VB12), this molecular monstrosity is a rearranger (isomerase), methyl extractor/dispenser, and halogen extractor. I need to add it but the initial rearrangement of AIR opens ribose like with histidine and Tryptophan, and the 1′ carbon is lost.
Tetra-Hydro-Folate, a methyl (-CH3), methylene (-CH2-), methenyl (-CH=), formyl (-CH=O), or formimino (-CH2=N) extractor/dispenser. This molecule is part of many 1 carbon processes. It dispenses the formate in purine biosynthesis. It’s involved with the transformation of serine to glycine and back. And it’s made from GTP. I’ll add it but the 5-membered ring of GTP is opened first between the nitrogens and the carbon between is lost as formate before ribose opens and all of the ribose is used in following molecules.
Lastly I added a part of Sulfur metabolism, the Iron-Sulfur cluster part that provides the sulfur in the Non-PRPP part of Thiamine biosynthesis in the form of carrier proteins being the sulfur where it is needed. Sulfur chemistry is a big part of this, and yet it’s not very directly involved in nucleic acid biosynthesis. It is critical in cofactors used to move carbon around (Thiamine, Biotin, coenzyme-A…).
PurF has what may be a vestigial iron-sulfur cluster though. Structural Biology of the Purine Biosynthetic Pathway: “The first type, present in higher vertebrates, plants, flies, cyanobacteria, and Gram-positive bacteria, contains a structural Fe4S4 cluster and a cleavable N-terminal propeptide, and is exemplified by Bacillus subtilis PurF in literature [10,16,17].”
Plans.
Add info about proteins and nucleotides as adaptors and dispensers/extractors of other molecules.
Add info about thematic differences among NTPs as P or PP donors.
Add more AARS (aminoacyl tRNA synthetase) info.
Attempt to organize the relationships by intermeadiates, and protein subdomain.
Add the urea cycle due to the connections between Purine (Asp amination), Pyrimidine (CP) and Arginine byosynthesis.
Add more reaction intermediates like aminoacyl-AMP. For example the glycine in purine biosynthesis exists as phospho-glycine. The aspartate as phospho-aspartate.
Other Observations.
Molecular “dispenser/extractor” and “adaptor”.
ATP, THF, glutamate, and other things ultimately may be providing things that were primordially available. So to an extent they, and some protein subdomains of associated enzymes, can be excluded from the picture. In fact aspartate and even glycine itself are ammonia donors in transamination reactions.
Similarly the same exclusion of parts can be done for molecules that act as adaptors, even tRNAs. I have ideas about coenzyme-A without the Adenosine-phosphate used to carry it around the cell.
Ideas.
Pre-cellular life. When looking at the NMPs and NDPs as adaptors the Pyrimidines have seem to have a bias towards membrane components and systems. CDP-glycerol and UDP and this part of bacterial cell wall biosynthesis.
G3P>E4P>R5P progression in backbone = each nucleotide in codon triplet?
Lysine is the only aa with a C1 and C1 AARS. Connection to Lysine looping steps in Lys biosynthesis via symmetry at the codon stem?
Glycinamide and bicarbonate, ammonia, or hydrogen sulfide as significant to abiogenesis.
Ribose originally come from a 3C+2C source? G3P + the activated aldehyde on Thiamine?
Issues.
The medium allows a never-ending cycle of reorganizing by different principals. On one level this is good. Copy and past my way to unpacking metabolism.
Good and Bad. All the things in my visual field is a potential benefit with ADHD. There are drawbacks.
Resources and how I use them.
Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology. 2nd ed. Gerhard Michael and Dietmar Schomburg, 2012. General reference.
Roche free molecular and cell biology posters. Sometimes I like to just stare at the whole mess.
Uniprot. This is a resource for protein sequence and functional information. This was my first stop in getting information about a protein, example PurF. After getting length and other info I scroll down to family and domain databases and check the Interpro links.
Interpro. This is a resource for classifying proteins into families and predicting domains and other important sites. Here’s PurF again. Scrolling down to entry matches for the protein you can see the what domains have been defined so far, 2 subdomains. Once I have the subdomains I go to ECOD.

ECOD. This is a resource that is a “… hierarchical classification of protein domains according to their evolutionary relationship.” Proteins aren’t just related to one another, their subparts are related to one another. Often separately. There are 4 levels of comparison X: possible homology level, H: homology level (big families), T: topology level (structural similarity), F: Families.
The first domain of PurF is in the alpha helixes + beta sheets 4-layers group. X: Ntn/PP2C (N-terminal nucleophile aminohydrolases/Protein serine/throinine phosphatases 2C), H: Ntn, T: Class 2 glutamine amidotransferases, F: GATase_2_1st. You can click right into it.
The second domain of PurF is in the alpha helix/beta sheet 3-layered sandwich group. X: PRTase-like (PhosphoRibosyl-Transferase), H: PRTase-like, T: PRTase-like, F: Pribosyltran.
With these 2 domains and boxes of their close relatives I can look for patterns that suggest general functions of the group, and how they relate to one another. There are many studies looking at relatedness among for example ATPgrasp enzymes and their subdomains.
And with all of these relationships in ribose metabolism visible on the diagram lots of patterns may become visible. It’s like a Rubik’s Cube/Matryoska Doll/Where’s Waldo combo. There are probably multiple Waldo’s.
KEGG. Sometimes it’s useful to look at more specific metabolic pathways. This is also good for finding everything a molecule like uracil might be involved in.
Pubmed. A major archive of journal articles.
Wikipedia. Often a good place to get general info.