I’ve been guilty of teaching bean-bag genetics this semester. Bean-bag genetics treats individuals as a bag of irrelevant shape containing a collection of alleles (the “beans”) that are sorted and disseminated by the rules of Mendel, and at its worst, assigns one trait to one allele; it’s highly unrealistic. In my defense, it was necessary — first-year students struggle enough with the basic logic of elementary transmission genetics without adding great complications — and of course, in some contexts, such as population genetics, it is a useful simplification. It’s just anathema to anyone more interested in the physiological and developmental side of genetics.
The heart of the problem is that it ignores the issue of translating genotype into phenotype. If you’ve ever had a basic genetics course, it’s quite common to have been taught only one concept about the phenotype problem: that an allele is either dominant, in which case it is expressed as the phenotype, or it’s recessive, in which case it is completely ignored unless it’s the only allele present. This idea is so 19th century — it’s an approximation made in the complete absence of any knowledge of the nature of genes.
And the “one gene, one trait” model violates everything we do know about the phenotype and genotype. Every gene is pleiotropic — it influences multiple traits to varying degrees. Every trait is multigenic — multiple genes contribute to the expression of every phenotypic detail. The bean-bag model is totally inadequate for describing the relationship of genes to physiology and morphology. Instead of a bean-bag, I prefer to think of the genome as comparable to a power spectrum, an expression of the organism in a completely different domain. But I wrote about that previously, and I’ll make this explanation a little simpler.
Here’s the problem: you can’t always reliably predict the phenotype from the genotype. We have a skewed perspective on the problem, because historically, genetics has first searched for strong phenotypes, and then gone looking for the genetic cause. We’ve been effectively blind to many subtle phenotypic effects, simply because we don’t know how to find them. When we go the other way, and start by mutating known genes and then looking for changes in the phenotype, we’re often surprised to discover no detectable change. One of the classic examples is the work of Elkins (1990), who found that mutating a neural cell adhesion gene, Fasciclin I, did not generate any gross defects. Mutating another gene, a signal transduction gene called Abelson tyrosine kinase, similarly had no visible effects. Mutating the two together, though — and this is a major clue to how these strange absences of effect could work — did produce gross and obvious effects on nervous system development.
Providing another great example, Steve Pinker examined his own genome, and discovered that his genes said he was predisposed to be red-haired and at high risk for baldness. If you’ve seen Steve Pinker, you know he’s neither.
How can this be? As any geneticist will tell you, the background — the other alleles present in the organism — are important in defining the pattern of expression of a specific gene of interest. One simple possibility is that the genome contains redundancy: that a trait such as adhesion of axons in the nervous system or the amount of hair on the head can be the product of multiple genes, each doing pretty much the same thing, so knocking out one doesn’t have a strong effect, because there is a backup present.
So Steve Pinker could have seen that he has a defective Gene A, which is important in regulating hair, but maybe there’s another Gene B lurking in the system that we haven’t characterized yet, and which can compensate for a missing Gene A, and he has a particularly strong form of it. One explanation for a variable association between an allele and the phenotype, then, is that we simply don’t have all the information about the multigenic cause of the phenotype, and there are other genes that can contribute.
This doesn’t explain all of the observed phenomena, however. Identical twins who share the same complement of alleles also exhibit variability in the phenotype; we also have isogenic animal lines, where every individual has the same genetic complement, and they also show variability in phenotype. This is the problem of penetrance; penetrance is a genetics term that refers to the likelihood that an individual carrying an allele will actually express the phenotype associated with that allele…and it’s not always 100%.
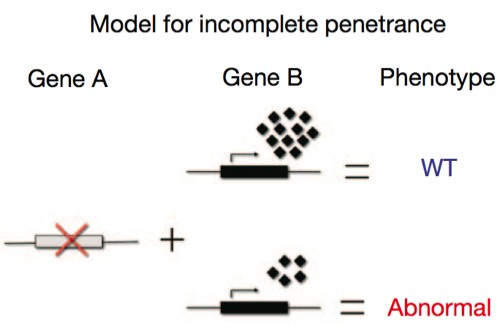
Again, the explanation lies in the other genes present in the organism. No gene functions all by itself; its expression is dependent on a cloud of other proteins — transcription factors, enhancers, chaperones — all of which modulate the gene of interest. We also have to deal with statistical variation in the degree of expression of all those modulatory factors, which vary by chance from cell to cell, and so the actual degree of activation of a gene may follow a kind of bell curve distribution. In the cartoon below, the little diamonds represent these partners; sometimes, just by chance, they’ll be present in sufficiently high numbers to boost Gene B’s output enough to fully compensate for a defective Gene A; in other cases, just by chance, they’re too low in concentration to adequately compensate for the absence.
What the above cartoon illustrates is the concept of developmental noise, the idea that the cumulative total of statistical variation in gene expression during development can produce significant phenotypic variation in the absence of any differences in the genotype. Developmental noise is a phrase bruited about quite a bit, and there’s good reason to think it’s valid: we can see quantitative variation in gene expression with molecular techniques, for instance. But at the same time we have other concepts, like redundancy and canalization, that work to buffer variation and produce reliable outputs from developmental processes, so we don’t have many good examples where we can directly correlate subtle variation at the molecular level with clear morphological differences.
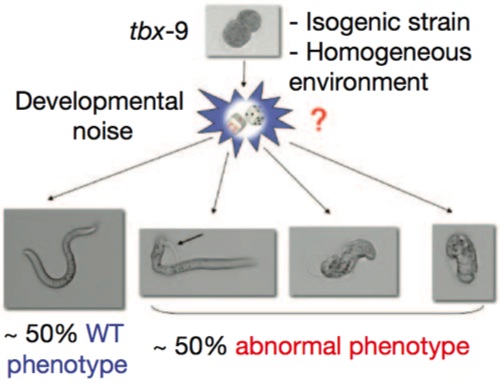
To test that, we have to go to simple animal models (it turns out that Steve Pinker is a rather intractable experimental animal). And here we have a very nice example in the nematode worm, C. elegans. In these experiments, the investigators were dealing with an isogenic strain — the genetic background was identical in all of the animals — raised in a uniform environment. They were looking at a mutant in the gene tbf9, which causes defects in muscle formation, but only 50% penetrance; that is, half the time, the mutants appeared completely normal, and the other half of the time they had grossly abnormal muscle development.
See the big red question mark? That’s the big question: can we trace the abnormal phenotype all the way back to random fluctuations in the expression of other genes in the animal? Yes, they can, otherwise it would never have been published in Nature and I wouldn’t be writing about it now.
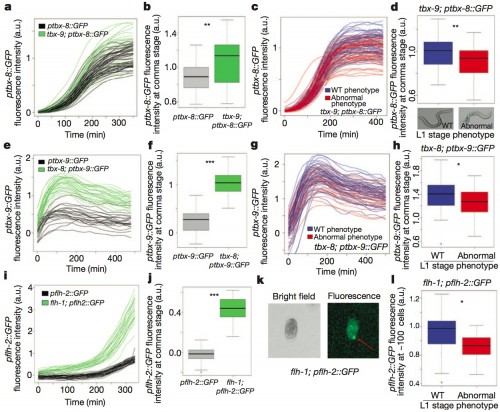
In this case, they have a situation analogous to the Gene A/Gene B cartoons above. Gene B is tbx-9; Gene B is a related gene, a duplicate called tbx-8 which acts as a redundant copy. In the experiments below, they knock out tbx-9 with a mutation, and then measure the quantity of other genes in the system using a very precise technique of quantitative fluorescence. Below, I’ve reproduced the entirety of their summary figure, because it is awesome — I just love the idea of being able to count the number of molecules expressed in a developing system. In order to avoid overwhelming everyone, though, I’ll just describe a couple of the panels to give you the gist of the work.
First, just look at the top left panel, a. It’s a plot of the level of expression of the tbx-8 gene over time, where each line in the plot is a different animal. The lines in black are in the wild type animal, with fully functional copies of bothe tbx-8 and tbx-9, and you should be able to see that there’s a fair amount of variation in expression, about two-fold, in different individuals. The lines in green are from animals mutant for tbx-9; it’s messy, but statistically what happens when tbx-9 is knocked out, more tbx-8 gene product is produced.
Panel e, just below it, shows the complementary experiment: the expression of tbx-9 is shown for both wild type (black) and animals with tbx-8 knocked out. Here, the difference is very clear: tbx-9 levels are greatly elevated in the absence of tbx-8. This shows that tbx-8 and tbx-9 are actually tied together in a regulatory relationship where levels of one rise in response to reduced levels of the other, and vice versa.
(Click for larger image)
Early inter-individual variation in the induction of ancestral gene duplicates predicts the outcome of inherited mutations. a, Quantification of total green fluorescent protein (GFP) expression from a tbx-8 reporter during embryonic development in WT (black) and tbx-9(ok2473) (green) individuals. Each individual is a separate line. a.u., Arbitrary units. b, Boxplot of tbx-8 reporter expression (a) showing 1.2-fold upregulation in a tbx-9 mutant at comma stage (~290 min, P=1.6×3 10-3, Wilcoxon rank test). c, Expression of tbx-8 reporter in a tbx-9(ok2473) background for embryos that hatch with (red) or without (blue, WT) a morphological defect. d, Boxplot of c showing tbx-8 expression is higher in tbx-9 embryos that develop a WT phenotype (blue) compared with those that develop an abnormal (red) phenotype at comma stage (P= 6.1×10-3). e, Expression of a ptbx-9::GFP reporter in WT (black) and tbx-8(ok656) mutant (green). f, Boxplot of tbx-9 reporter showing 4.3-fold upregulation at comma stage (~375 min, P=3.6×10-16). g, Expression of tbx-9 reporter in a tbx-8(ok656) mutant background, colour code as in c. h, Boxplot of g showing tbx-9 expression is higher in tbx-8 embryos that develop a WT phenotype (P=0.033). i, Expression of a pflh-2::GFP reporter in WT (black) and flh-1(bc374) mutant (green). j, Boxplot of flh-2 reporter expression (i) showing 1.8-fold upregulation in a flh-1 mutant at comma stage (~180 min, P=2.2×10-16). k, Bright-field and fluorescence image of an approximate 100-cell flh-1; pflh-2::GFP embryo. Red arrow indicates the local expression of flh-2 reporter quantified for flh-1 phenotypic prediction.
l, Boxplot showing higher flh-2 reporter expression at approximate 100 cells for WT (blue) compared with abnormal (red) phenotypes (P=0.014). Boxplots show the median, quartiles, maximum and minimum expression in each data set.
Now skip over to the right, to panel c. All of the lines in this plot are of tbx-8 expression in tbx-9 mutants, and again you see a wide variation in levels of gene expression. In addition, the lines are color-coded by whether the worm developed normally (blue), or had the mutant phenotype (red). The answer: worms with low tbx-8 levels were more likely to have the abnormal phenotype than those with high levels.
Panel g, just below it, is the complementary analysis of tbx-9 levels in tbx-8 mutants, and it gives the same answer.
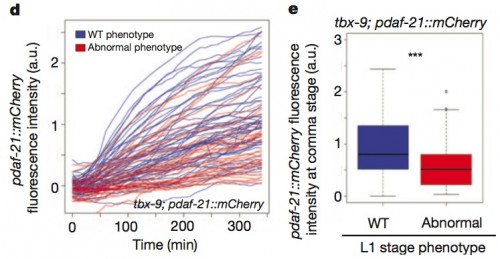
Obviously, though, there is still a lot of variability unaccounted for; having relatively high levels of one or the other of the tbx genes didn’t automatically mean the worm developed a wild-type phenotype. There’s got to be something more that is varying. Look way back to the second cartoon I showed, with the little diamonds representing the cloud of transcription factors and chaperone proteins that modulate gene expression. Could there also be correlated variation there? And yes, there is. The authors looked at a chaperone protein called daf-21 that is associated with the tbx system, and found, in mutants for tbx-9, that elevated levels of daf-21 were associated with wildtype morphology (in blue), while lowered levels of daf-21 were associated with the mutant phenotype.
(Click for larger image)
Expression of daf-21 reporter in a tbx-9(ok2473) mutant background. Embryos that hatch into phenotypically WT worms (blue) have higher expression than those hatching with a morphological defect (red) at the comma stage (P=1.9×10-3).
I know what you’re thinking: there isn’t a perfect correlation between high daf-21 levels and wildtype morphology either. But when they do double-label experiments, and take into account both daf-21 and tbx-8 levels in tbx-9 mutants, they found that 92% of the animals with greater than median levels of expression of both daf-21 and tbx-8 had wildtype morphology. It’s still not perfect, but it’s pretty darned good, and besides, it’s no surprise that there are probably other modulatory factors with statistical variation lurking in the system.
What should you learn from this? Developmental noise is real, and is a product of statistical variation in the degree of expression of multiple genetic components that contribute to a phenotype. We can measure that molecular variation in living, developing systems and correlate it phenotypic outcomes. None of this is surprising; we expect that the process of gene expression is going to be a bit noisy, especially in these transcriptional regulators that are present in low concentration in the cell, anyway. But the other cool thing we can observe here is that having multiple noisy systems that interact with each other can produce a more reliable, robust signal and contribute to the fidelity of developmental outcomes.
Burga A, Casanueva MO, Lehner B (2011) Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature 480(7376):250-3.
Elkins T, Zinn K, McAllister L, Hoffmann FM, Goodman CS (1990) Genetic analysis of a Drosophila neural cell adhesion molecule: interaction of fasciclin I and Abelson tyrosine kinase mutations. Cell 60(4):565-75.
(Also on Sb)



Pinker is in fact a member – moreover, the first member – of the Luxuriant Flowing Hair Club for Scientists.
The intense study continues.
Dunno if you’re aware of this, but Tom Elkins–first author of the paper from Goodman’s lab on Fas-Abl interaction–died before finishing his PhD, killed by a drunk driver. There is a biennial award in his memory for outstanding graduate student research in Drosophila neurobiology:
http://meetings.cshl.edu/meetings/elkins_solicitation_11.pdf
Bean bags are so out of style by now.
Get some fashion sense, geneticists.
Glen Davidson
As someone who was really interested in biology in high school, to the point of almost wanting to go into that field, but is now an engineering major, I find this very interesting. It’s nice to have a simple way to explain these thing beyond the “You can only have blue eyes if you have recessive genes” conversation. And I’ll admit, I’m a sucker for pictures in science articles.
What are the odds that some fundie will (ab)use the introductory part of PZ:s post in order to claim that ‘even scientists admit science/genetics/biology is all wrong’?
Yes, I knew Elkins — not well, but we were both grad students in dev. neurosci. at the same time, at different institutions. I remember being shocked when I learned about his death.
Sorry, I need to correct my earlier comment: Someone has pointed out to me that Tom Elkins actually had finished his PhD before he was killed, and was in the early stages of his post-doc training in Goodman’s lab.
This reminds me of when I learned that the “solar system” model for atoms was wrong.
The complexities with human genetics makes me wonder if we’re going to get Cyborgs before Genetically Engineered People.
Steven Pinker is a “borrowed ladder” – PZ is just covering for him with all this diversionary genetics…
Interesting science and conclusions. Not really surprising when one thinks about it.
Never heard of a developmental biologist accused of oversimplifying anything.
Amazing stuff, PZ. I wish you’d write more about stuff like this; you do a great job of explaining what’s going on. I’ll bet if I had read the article, I wouldn’t know what the hell I was reading.. but I was able to follow your post without a problem.
Meanwhile, the creationists are still asking why there are still monkeys if we’re evolved from monkeys.
“In order to avoid overwhelming everyone, though,….”
Sheesh!
In one physics class, my professor once talked about ‘perfectly-spherical cows’, which is a catchall term for any problem or example meant to remove excess complications from a problem. So, while you may not like a bean-bag cow filled with alleles, at least you weren’t talking about perfectly spherical, completely homogeneous cows.
Bean bag genetics was the standard presentation to beginners at least up until ca. 1960.
There’s nothing wrong with an introductory course in genetics using that approximation as long as they are repeatedly told “This course is a simplification, an approximation; the real situation is considerably more complicated. So don’t think that the world is entirely about simple Mendelian recessive-dominant genes that each control one macro-feature of an organism.”
In fact, the bean bag approach is probably very good pedagogy as it allows you to take a historical approach by outlining Mendel’s observations and his conclusions about the underlying mechanism. As time permits you can then progress chronologically and bring up one by one significant observations that cannot be explained by the Mendelian model.
Teaching science of any kind is extremely difficult. It’s a subject I’ve given much thought to, concluding that in introductory courses a historical approach may be best. In my own field, chemistry, introductory texts present more or less our current state of understanding, and the hapless student does not realize that the development of that understanding took a lot of hard work by a lot of very smart people over a very long time. Bringing in a historical perspective allows the students’ understanding to develop along the same lines as did that of the chemists, incrementally, one piece at a time, instead of expecting the student to swallow the whole thing at once, holus bolus.
As chemistry, so genetics, though the time frame for the development of genetics is considerably shorter.
Carry on, P-zed. Steady as she goes.
The first two links in the article are to Haldane & Crow defending bean-bag genetics, and I’m not going to gainsay those two.
Bad ass.
I remember learning bean bag genetics. Of course, that’s about where my genetics education left off. There’s nothing wrong with (and it’s generally a sound pedagogical approach) teaching simplified models to start with, to get concepts across to those who are new to a subject.
I’ve seen people argue that multiplication is not just repeated addition and it should never be taught that way. However, after a number of years of tutoring math from basic arithmetic through Calc and Diff EQs, I’ve found it to be a good learning model, even if it’s not 100% accurate at higher levels.
Keep on keepin’ on.
Nice discussion, P.Z.
For anyone interested in an easily digestible review of the “bean bag genetics” debate between Haldane and Mayr, I would suggest taking a look at this article from the Journal of History of Biology.
J. B. S. Haldane, Ernst Mayr and the Beanbag Genetics Dispute
J Hist Biol
2010 vol. 44 (2) pp. 233-281
Anything worth learning is never simple. But one has to start with the simple things to have the necessary background to understand the more complicated things.
I love your research blogs, PZ. They simplify my job when I need to explain genetics to the non-geneticist or non-molecular biologist. You are always able to explain it so clearly. I wish you would do research blogging more often!
I learned in school that there was also partial dominance, groups of genes working together, and incomplete penetrance. Later, I read something even more interesting: that certain mountain frogs have different genes that, operating at different altitudes, give them the same phenotype! And then there are mere developmental variations. I love it.
Anyway, it’s “one gene, one peptide chain,” isn’t it?
With alternative splicing, it’s more like “one gene, one or more peptide chains.”
And then of course genes that code for functional RNA instead of peptides.
I’m trying to follow, I really am. Its nice of you to simplify it for us PZ. But I keep getting distracted every time you say “mutant”. I keep wondering which ones get the accelerated healing and which ones get the laser eyes.
So full of win!
Awesome. I’ve just finished Stephen Nowicki’s course on Intro Biology from the Teaching Company. I can definitely say that I understood a whole lot more of that post than I would have a month ago. I certainly understood the initial phenotype/genotype problem immediately. I might have to parse this post once or twice more for completeness. I am after all merely an engineer.
That parenthetical sentence evoked a mental image of Pinker crouching in the corner of a cage surrounded by geneticists in lab coats. Pinker is clutching his flowing locks, sticking out his tongue and giving the scientists a big raspberry and chanting, “Nya nyanyaa!”
PZ’s got luxuriant flowing hair, it just flows from the lower half of his face.
Excellent. Just 3 more years of Pharyngula and I can get my honorary degree in Atheism with a sub-major in Biochemistry and Genetics.
Great article, PZ. It seems as if I’m not the only one who would be interested in more of these biology-based blogs.
This article also came as a surprise to me as I have been taught only about dominant and recessive alleles; though to be fair I was taught this in Biological Anthropology courses and not strictly biology courses.
aa: Thanks for the Rao and Nanjundiah reference. Cool paper. The chummy fistiness of Haldane and Mayr’s friendship is much more common in science than, you know…not in science. At least in my experience.
Finally an appropriate banner ad!!! Someone sells micro-RNA inhibitors.
“Do you know what your micro-RNA is doing?”
Sorry about the hyphen.
“Click here to see the recent microRNA microarray comparison”
“Can you detect ALL microRNAs?
Regardless of GC content”
Capitalism for nerds! :-)
Or as T’Perry called it Lies to children.
If Martyn Poliakoff is not a member of that club, the whole concept is a joke.
Arrgh. Its P’Terry (Terry Pratchett). Now where did I leave my dried frog pills?
Re CSMiller and his confusion between T’Perry and P’Terry:
Does it help any to know that the name of the capital of Georgia, Tbilisi, is derived from an earlier word t’pilisi, referring to the hot springs there? IOW, you are not alone. Almost, but not entirely.
Georgian includes the word გვფრცქვნის (gvprtskvnis), with the technical meaning “He is peeling us” but idiomatically meaning “he is ripping us off.” How the Georgians manage a long strings of consonants like this is beyond me.
T’Perry indeed.
I remember learning simple mendelian genetics, and at the same time being told that very few human traits follow that pattern. Two that do, AFAIK, are the ability to roll your tongue up, and having detached earlobes.
I certainly agree that modern genetics should seek to elucidate the relation between genes. Still, I think what you characterize as bean bag genetics deserves much more respect. You must remember that what we call molecular biology today was solely built on this kind of genetic research. At least in phage/bacterial genetics it was definitely never the case that interactions of different alleles or genes have not been paid attention to. Quite the opposite: It was the interaction of different alleles/genes that led to some of the biggest intellectual achievements in biology. E.g., identification of DNA as the genetic material, the central dogma of molecular biology, the discovery of the genetic code and the operon model. And all this without knowing much about the nature of the gene. Those who are interestes should read Judson’s “The eighth day of creation”.
In addition, all knock outs in mice, IMO the current standard in mammalian genetics, start as bean bag genetics, i.e. researchers look for a single phenotype in a given genetic background. Still, much of our understanding of mammalian genetics/geneomics stems from conclusions derived from counting beans. Trying to compensate for or to enhance phenotypes through breeding with other lines most often follows educated guesses rather than real rationales but still lead to significant contributions to, e.g., interaction of signalling pathways. Still, for the vast majority of mammalian genes we currently don’t even such data and researchers are working to get hold of a bag of beans they could count.
On the other hand it’s true that no gene acts in a vacuum isolated from other genes and there are indeed nice publications on the interaction of genes. However, we currently observe that a tendency to declare that everything is linked to everything has come into vogue thereby, IMO, trivializng the issue. Often such publication are based on computer models rather than wet lab data (to a degree this may relate to the fact that people from mathematics/physics with limited knowledge in even bean bag genetics are attracted by field). As mentioned above much of the current mutant research at least in mice is closer to bean bag genetics than to what people suggest systems biology should be. This is due to the fact that most mutations are investigated in a single or a few genetic backgrounds in a stable environment. Thus, many if not most interactions of a single trait will just not become visible. There are efforts to overcome this situation by by the Colaborative Cross which will derive 1000 recombinant inbred lines from intercrosses of six different house mouse strains and two closely related species. However, working with these strains will be counting beans again but the very same method though on a smaller scale led to the discovery of Toll like receptors as pathogen receptors in mice and earned Bruce Beutler the Noble Prize for Medicien and Physiology. Thus, bean bag genetics is alive and still going strong.
Those graphs kinda look like Pinker’s hair.
or worms.
Sir:
I’m a fairly smart person, scientifically educated at a technical university, and vaguely interested in how the world works and what makes things tick. I was taught (in the 1970s) the usual dominant/recessive Mendelian simplification. Later in life, I learned that things are (of course) more complex, and while I was unsurprised about this, I didn’t know much in the way of detail.
This essay blew my socks off. This is awesome stuff. You did nothing more than explain, from a fairly high level, what was going on with the genetic dynamics in Stephen Pinker and a population of nematodes. It wasn’t even your work; you’re just narrating a paper from Nature, basically. But there is no way that I could have followed and understood the Nature article, let alone understood it in terms of the broader topic of gene interdependence. That, you provided.
Excellent, sir, excellent. And thank you very much.