Somehow this game hit a spot for me, and I want to dissect it. Not the spot since I’m still looking for the perfect something on Android (if it featured some match 3 play that would help). But it’s nice overall, something about helping other people fight monsters. But the formula breaks down a bit when I think about it, or overthink it as some might say. But I think it’s valid to look at games like this even while enjoying them.
Dungeon Village 2 is a town building game with a town space and an outside battle space.

As you improve your village more people are attracted to it and go outside to fight monsters. They all get the traditional RPG improvement mechanics where they get experience and gold, level up, and buy themselves equipment in the weapon, armor, and accessory shops in the village. You can gift them equipment which is much more efficient. Then they go back out and do it some more. They also get knocked out instead of dying and becoming coins for the monsters, and will rush one another to the inn to rest up. Bosses will also show up periodically.
You get to build the town, including houses for people who become citizens instead of visitors (and they pay taxes). Hire people for quests (dungeons or concentrations of monsters), meet requirements for upgrading towns, hold special events that increase stats or town popularity, as well as meet requirements for contests and special town visitors (they spend more money mostly).
The monsters are sentient, they talk. This is where things get weird for me. There’s this unstated history of conflict where these creatures are constantly fighting towns and keep coming back. Why?
If you have a monster tamer in a group for a quest there’s a chance a monster will be impressed with you defeating them and ask to join you. You can assign them to a fighter who forms a “partnership” (suggesting a kind of equal status) with the monster and they then go out together to fight the former allies of the monster. When the monster’s happiness is high enough it lets the fighter ride it into battle. Monsters without a partner live at the monster farm. I guess monster civilization isn’t good.
Speaking of monster civilization what’s with the gold you get? This suggests monster economics upon defeat, or some kind of evolution where flesh becomes metal on death. Strange stuff.
While this could be a bit overthinky to this point (I do wonder about what we feed into culturally though) the next bit definitely isn’t. Trigger warning for racism.
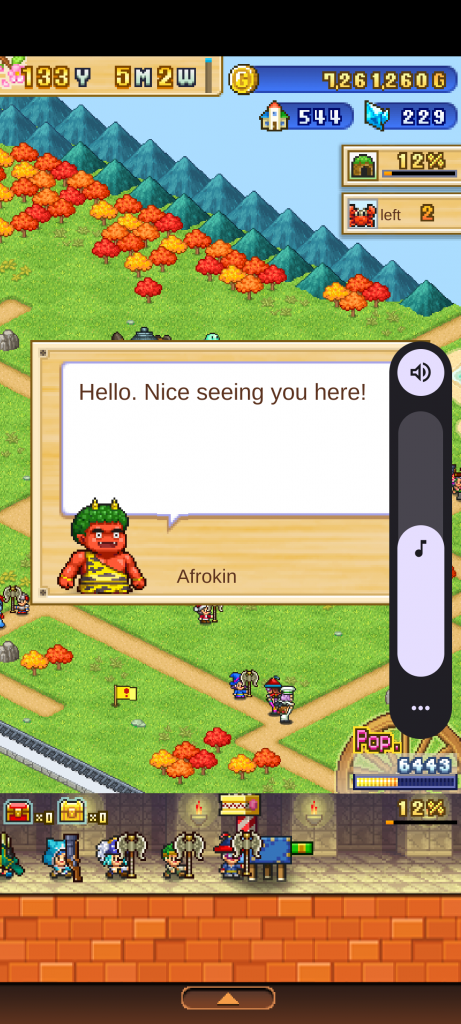
Sorry for the volume slider but this is the boss Afrokin. An apparent demonesque humanoid wearing animal skins with an afro. This is an insulting connection between people who have afros (people of African descent) and apparent barbarity and being a monster.
After the character says the above dialog they say “Let’s get down to business. I’m going to give you a painful lesson in fashion sense. My pants are stylish, comfortable, superior! There’s no way I’m going to lose while I’m wearing these!” When they lose they say “Not bad but the way I see it I only lost to myself, not you!”
This is all sorts of disturbing and doesn’t belong in any video game.
When I’m done with this game I’d like to find something similar on Android. Flawed but fun outside of the racism and apparent beating of monsters into joining you instead of finding a way to live together without conflict. If any of you have a suggestion for me about a similar game on Android I’d love to see it.