I want to put all of those protein domains in the previous posts into a larger context. Some of this I mentioned in previous posts in this series but they are worth mentioning again. I post a part of the big drawing to start.
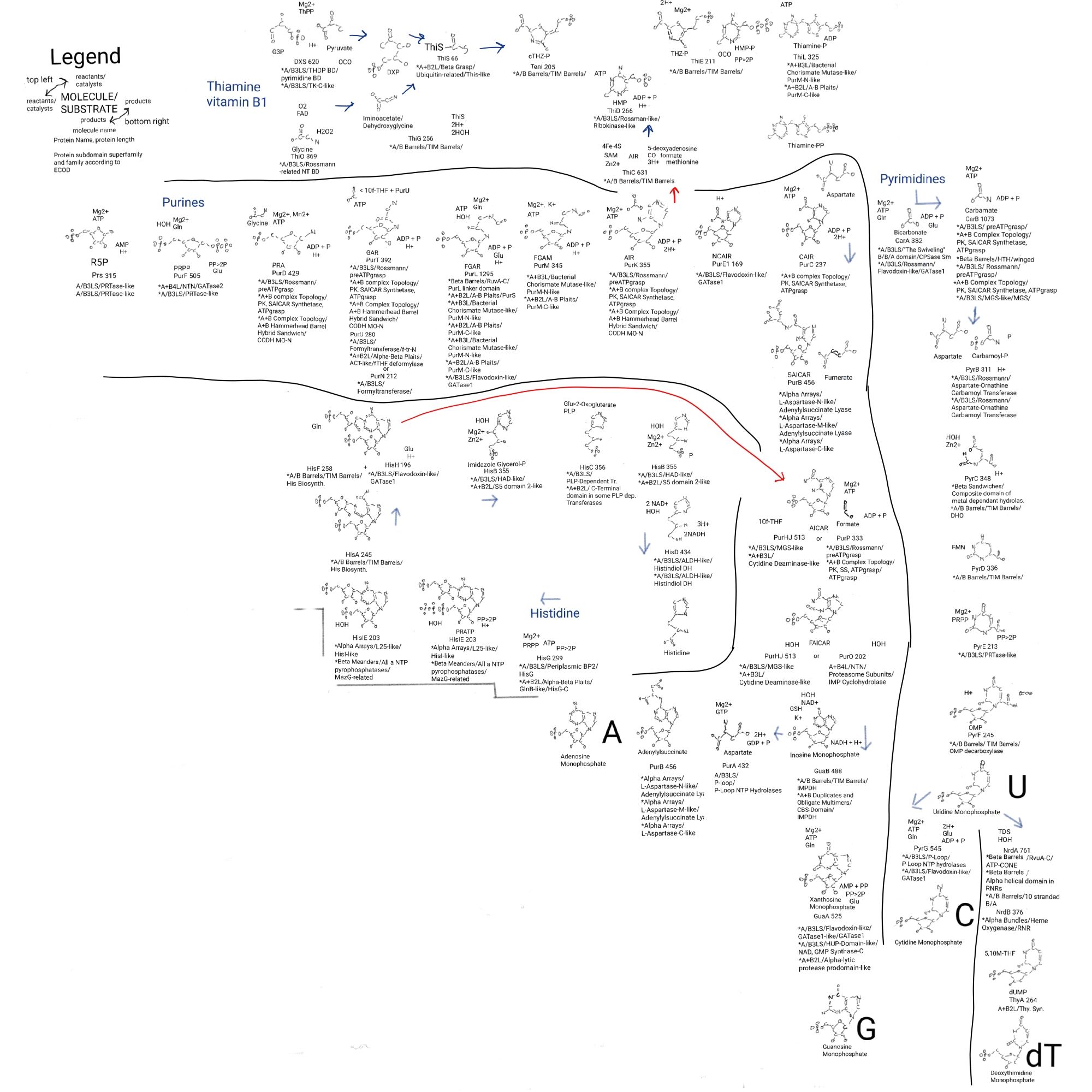
Nucleotides do a set of things. I hope I have covered them all and there aren’t any things they do that I missed.
- Nucleotides are made, in specific biochemical pathways, and separated into purine (AG) and pyrimidine (UCdT) pathways. These pathways merge with thiamine and histidine pathways.
- Nucleotides are genomes, messenger RNAs, ribosomal, and transfer RNAs (and other associated RNAs). They base pair through polar interactions between their carbonyls (C=O) and amines (-NH3/-N=).
- Nucleotides are the biosynthic source for other cofactors like vitamins. The output of purine biosynthesis becomes folate, flavin…
- Nucleotides are dispensers for phosphate, and not just ATP. There are thematic differences the kinds of things ATP, GTP, and CTP dispense for.
- Nucleotides are handles and escort molecules for cofactors including many vitamins, membrane components and some other molecules. And handles for amino acids during translation in tRNA.
- Nucleotides along with amino acids are the substance of transcription and translation. The base pairing becomes patterns of amino acid sequence that determine protein sequence.
- Nucleotides are signalling molecules, often when cyclized, alone and in multiples.
- Nucleotides act as replaceable groups like in sulfur transfer and Adenylylation.
Hydrothermal systems, minerals and ocean chemistry seem to somehow expand into this.
Nucleotide biosynthesis.
First there is the variable of the biochemical pathways themselves. What was the early extent of vent chemistry generated nucleotides? Did the reactions race straight to inosine or AMP? Or did things accumulate at points like the first ring closure? Or glycine binding? Did glycine binding repeat in an early polypeptie? Maybe there was a population of 5-membered pyrrole (5-membered carbon and nitrogen ring) ring creatures that could catalyze things towards inosine?
Looking at part of the big drawing again the similarities between thiamine thiazole biosynthesis and stage 1 of purine biosynthesis are interesting. A 5-carbon chain is made from a 2C and a 3C (DXP), and it is combined with a modified glycine and sulfur. And right after purine first ring closure that molecule can be made into the second part of thiamine, AIR becomes HMP. There’s even an imine on glycine in both cases (C=N). There’s this interesting imine on aspartate in niacin biosynthesis too.
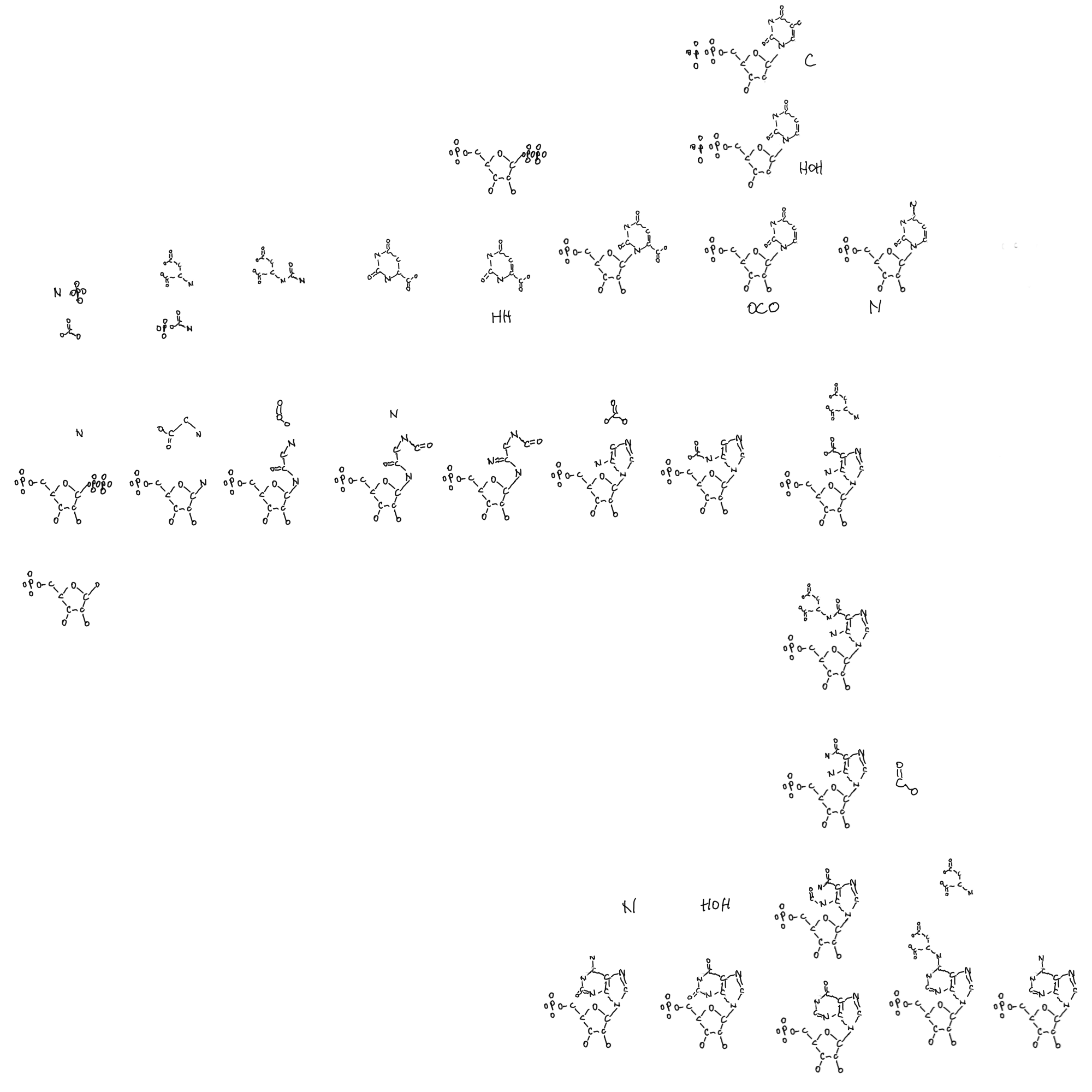
And where do the pyrimidines fit in with their ring synthesis and then attachment to ribose? Was there a point where pyrroles and pyridines (5- and 6-membered carbon and nitrogen ring) coexisted on ribose-phosphate chains? How does niacin fit in with ring creation and then ribose binding, and imine on aspartate?
And how do I think about the way the purines and pyrimidines have opposite relationships with ribose? Purines being built onto ribose and pyrimidines built and then put onto ribose. And niacin, NAD(P), built very much like pyrimidines, does that matter?
Genomes, RNA, and base pairing.
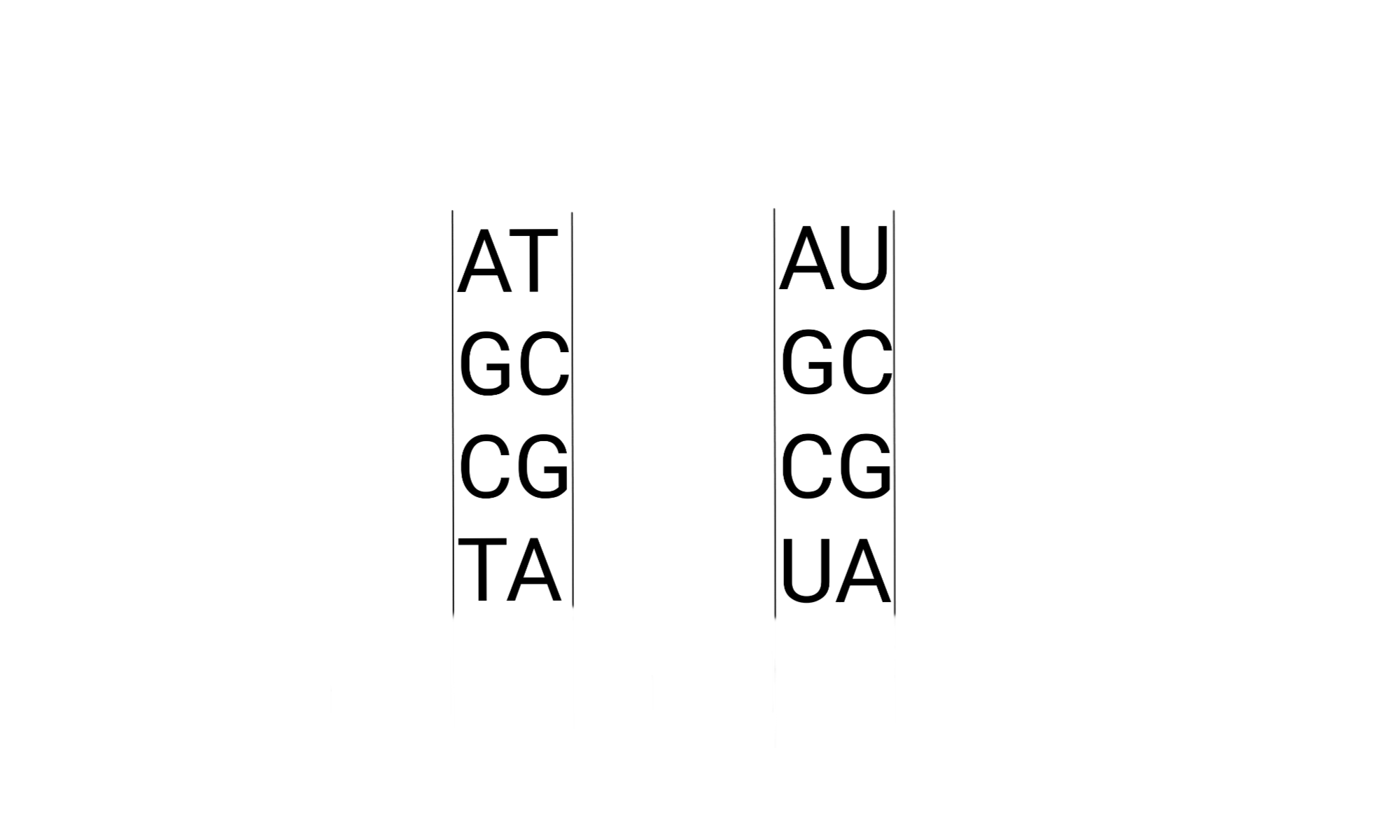
This involves interactions between strands of the same or different nucleotides. In DNA genomes and RNAs these are attractions based on polarity, but what about other interactions as one goes back? Can we count on today’s sequences being AGCU? (I believe it was once just RNA genomes) Maybe they blur down into base pairing between other 5 and 6 membered rings? Intermediates in nucleotide biosynthesis?
Was chemistry between strands a thing? Are genomes, transcription and translation just massive off-loading of functions from an ancestral molecule that did more than sit there as an information source analog? Did it make itself through chemistry it participated in as something that looked like a purine intermediate like AIR?
Nucleotides as biosynthic sources of amino acids and cofactors.
Nucleotides are the biosynthic source of the amino acid histidine, and cofactors like folate, riboflavin, and molybdenum cofactor. GTP is specifically what is dismantled for the cofactors, and ATP for histidine.
Maybe there are clues for the origin of ribose in how it is deconstructed in these things and tryptophan? PRPP is the ribose source in tryptophan, not nucleotides, but that’s part of the story too. After completing this more complicated drawing involving nucleotide metabolism I think about being able to peel back later evolving parts of metabolism and trying to find connections to more ancient parts.
Nucleotides as phosphate dispensers.
ATP is the stereotype for dispensing phosphate for active site chemistry and molecular movement. But GTP does it too and specifically for translation and cytoskeleton proteins like microfilaments and microtubules, things that could be useful for getting around in cells or mineral systems. CTP sometimes drives cysteine attachment to make coenzyme-A and it plays a phosphate dispenser role in cell division (ParB, chromosomal partitioning), hydrolysis indicating a completed step. Otherwise all of the NTPs and dNTPs can donate phosphate to one another in interconversions.
Nucleotides as handles.
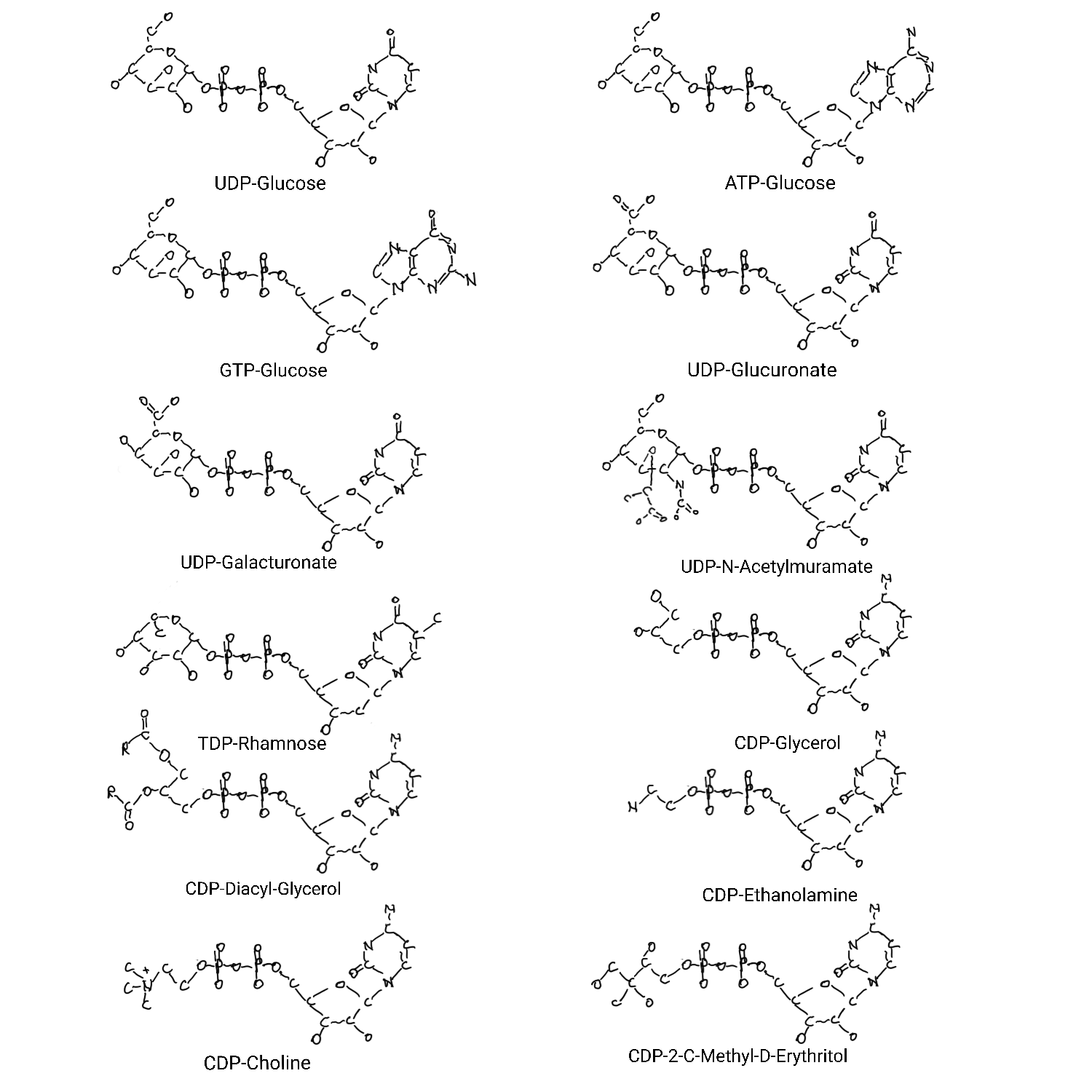
ATP acts as a handle for many cofactors like niacin, riboflavin, coenzyme-A, S-adenosyl-methionine, and thiamine. GTP and CTP acts like a handle for parts of molybdenum cofactor. All of the bases act as handles for kinds of carbohydrates ,UDP-glucose has the feel of a most ancient part, and CDP has a theme involving membrane components.
It’s interesting that deoxythimidine handles deoxycarbohydrates. A theme carried over. It makes me wonder about themes with ADP, UDP, and GDP as carbohydrate handles.
Nucleotides as the substance of transcription and translation.
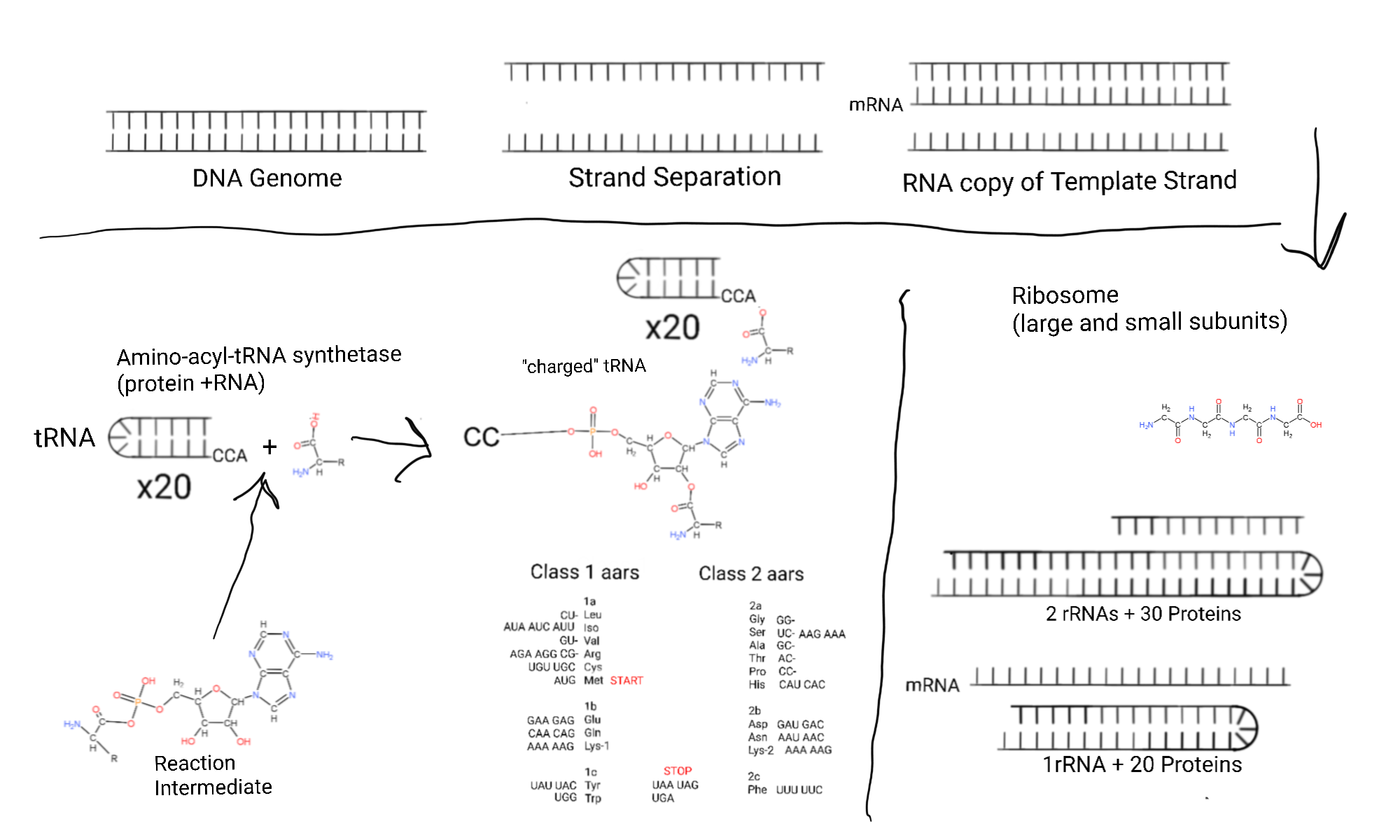
This is what biology class covers. The actualized genetic code. The process where base pairing in DNA and/or RNA becomes specific amino acids joined in a polypeptide chain. Like the protein subdomains I listed for every protein on the big drawing. tRNA is another way that nucleotides are handles, but for amino acids.
Maybe it all goes to polyglycine originally. Glycine is involved in making purines, the thiamine 5-membered thiazole ring, and every other amino acid looks like a modified glycine. Which leads to another question, could other amino acids have been made right on early polyglycine? Or were they made separately like today and added to a polypeptide with glycine?
Nucleotides as signalling molecules.
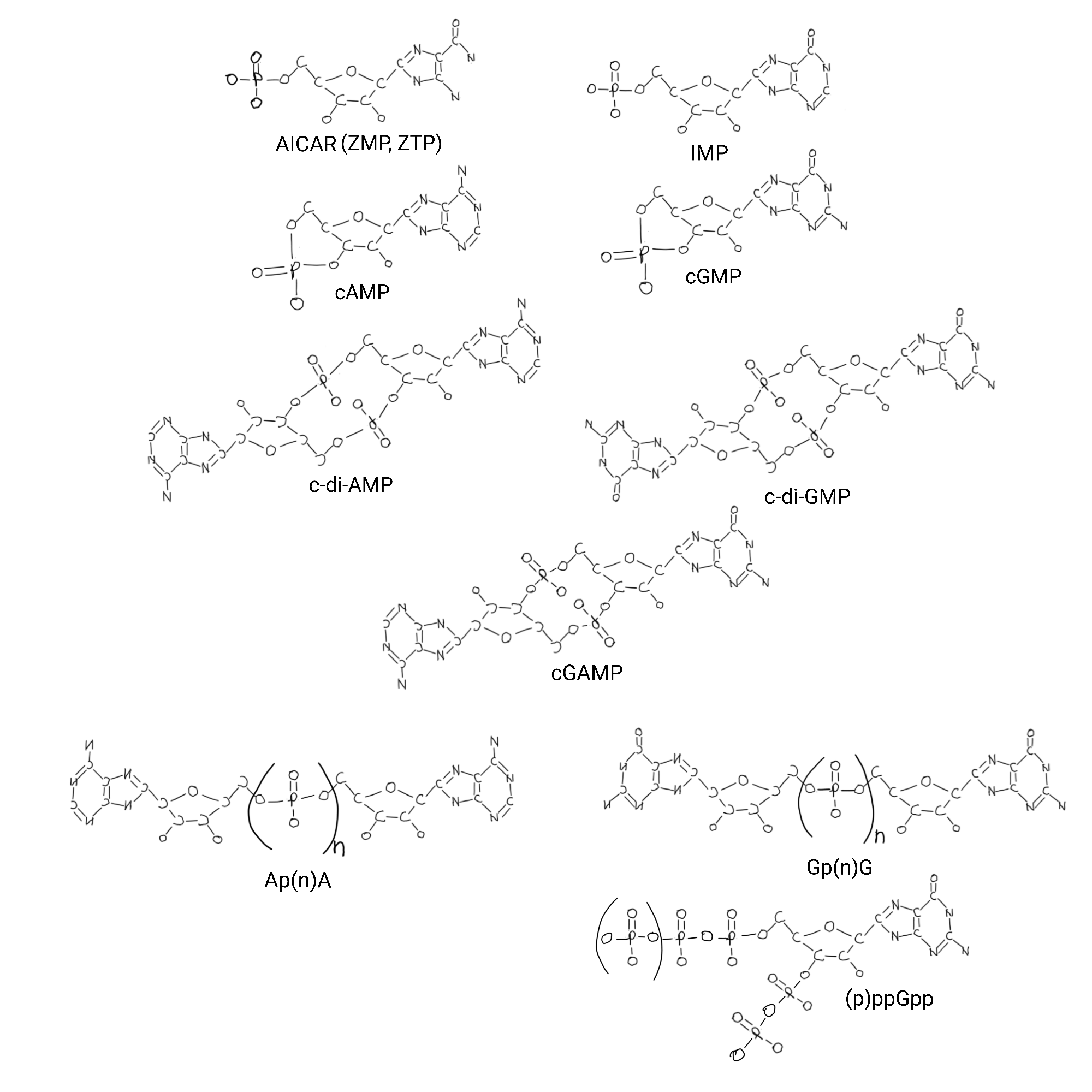
Nucleotides are part of rapid intracellular communication networks as cyclized forms. Single nucleotides can be cyclized at the phosphate, or cyclic dinucleotides connected at their phosphates. And other forms with extra phosphates in different places that act as signals inside of a cell.
All that phosphate. Proteins detect specific phosphates and numbers of phosphates in specific locations in so many ways I have no problem with the idea that early biology emphasized it in protein domains. Maybe phosphate was in the very first parts of selection.
Nucleotides as replaceable chemical groups.
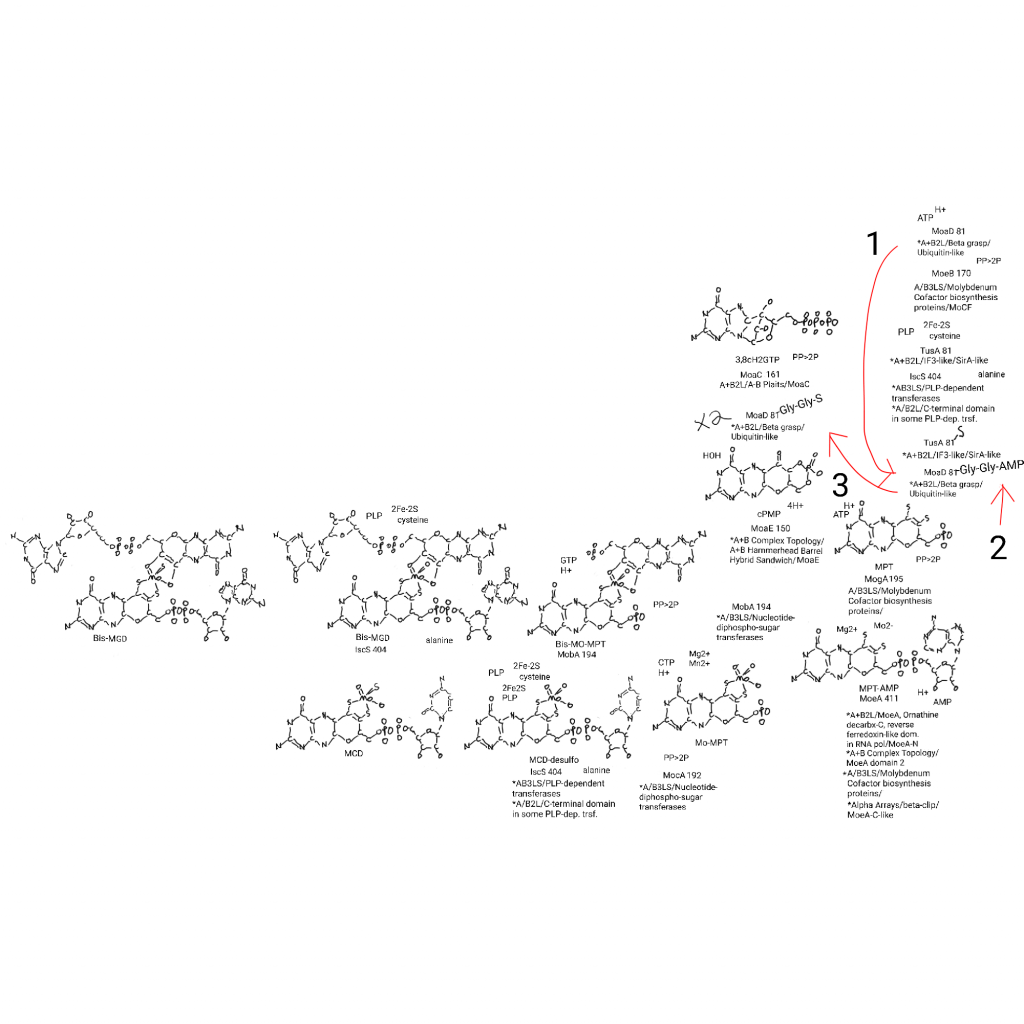
Here is the molybdenum cofactor part of the big drawing. Nucleotides are part of the logic of sulfur transfer to sulfur carrier proteins (red arrows and 1-3), here MoaD, 2 of which transfer sulfur to the molecule by replacing AMP (an adenylylation) added by MoaB. To a double glycine residue which is very interesting since I think things go to polyglycine somewhere in history. This also happens with ThiS in thiamine biosynthesis and I want to add that to the big drawing at some point.
This is an area that is largely unexplored by me like nucleotides as signalling molecules and nucleotides as handles outside of translation. It’s to the point where I may need a separate big drawing from everything nucleotides do in addition to the drawing that shows how they are made and what they are linked to in biosynthesis.
And that’s it for the tour of nucleotide biosynthesis, the proteins that carry it out, and their relationships with the rest of metabolism. I hope this has been interesting and others now find these origin of life puzzle pieces as interesting as I do. I may be short on answers but it’s about finding clues to how it may have happened. Being able to imagine how it could have looked given that we collectively probably won’t know for sure in our lifetimes. At least there is substance for the imagination.

It is not an exaggeration to assert that Five Nights At Freddy’s has earned the title of “best survival horror game” in recent times. If you are avidly interested in this genre, do not pass up the chance to experience it. I am confident that it is a battle royale game that is well worth playing.