Stage 1, formation of the first purine ring (a 5-membered C and N ring is a pyrrole) is not only complete, I have covered a bunch of protein families I can refer back to.
I did not expand on the relatives of one family that I will expand on in this post though. The preATPgrasp domain relatives are a group I wasn’t sure how I wanted to discuss yet.
Metabolism broadly.
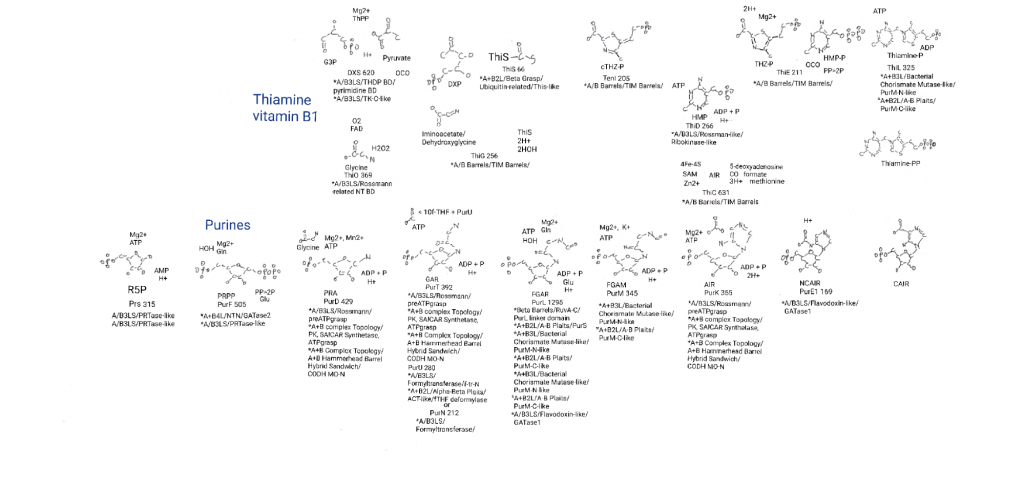
This is also a good point to mention that purine biosynthesis merges with 2 other metabolic pathways here. AIR can be made into the 6-membered ring (a pyridine) of thimine, HMP. What I did not put on the big drawing this time was that AIR can also be made into the “lower ligand” of vitamin B12. A ligand is an atom or molecule that binds to a molecule or protein to help carry out its function (the cyanide groups in the last post were ligands). B12 itself is a chelator of cobalt. But there are other lower ligands (potentially less origins related) and I was running out of space.
Stage 2: binding and movement of bicarbonate.
Life has 2 ways of doing the first part of stage 2 but I used the more complicated route because the direct binding of carbon dioxide to where bicarbonate moves to (by class 2 PurEs) is only in animals.
Stage 2.1: bicarbonate binding.
PurK is the protein that binds a bicarbonate to the nitrogen sticking out from the ring. And we have already seen these protein subdomains. It’s preATPgrasp, ATPgrasp, and Carbon monoxide dehydrogenase molybdoprotein N-like again. So for most of what is interesting I will refer to a previous post.
As an observation I would point out that all that is really needed to make these reactions work so far is phosphate. Every reaction but one so far uses a phosphate in active site chemistry, and the exception is the PurN/PurT step where life can add formate without phosphate, and yet PurT uses phosphate anyway in other organisms (E.coli has both).
What I will discuss this time are the relatives of the preATPgrasp domain. This fragment and its family members are in their own topology bin and there are 13 other topology siblings within a larger Rossman-related homology group, inside of a Rossman-like bin, inside of the A+B 3 layer sandwiches.
There are a lot of Rossman-related domains. If I had to stereotype the group based on the associations I would label them “nucleotide binders with a bias for Adenine containing cofactors”. But that is not based on structure, just looking in the bins and they don’t exclusively bind nucleotides.

The first 4 topology groups to mention are: a NAD(P) (niacin) binding Rossman-fold domain, a FAD (riboflavin)/NAD(P) binding domain, a DHS-like NAD/FAD binding domain, and a “Nucleotide binding domain” bin that seems to do all of the above. These are all electron/proton dispensers/acceptors, and the first 2 contain many families, too many to go over in detail. Glycosyltransferase Mag is flagella related so I left it out.
But there is a whole area of biochemistry related to “nucleotides as handles” that I saw inside of the above bins that I haven’t covered on the big drawing. You see Adenosine (adenine-ribose-phosphate) used as a handle for cofactors all over the big drawing, and some of the other bases are used as handles in the molybdenum cofactor part of the drawing. But all of the bases have jobs as handles and there is a range of things related to membranes and sugars that adenosine, guanosine, uridine, cytosine, and thymidine are involved in as handles.
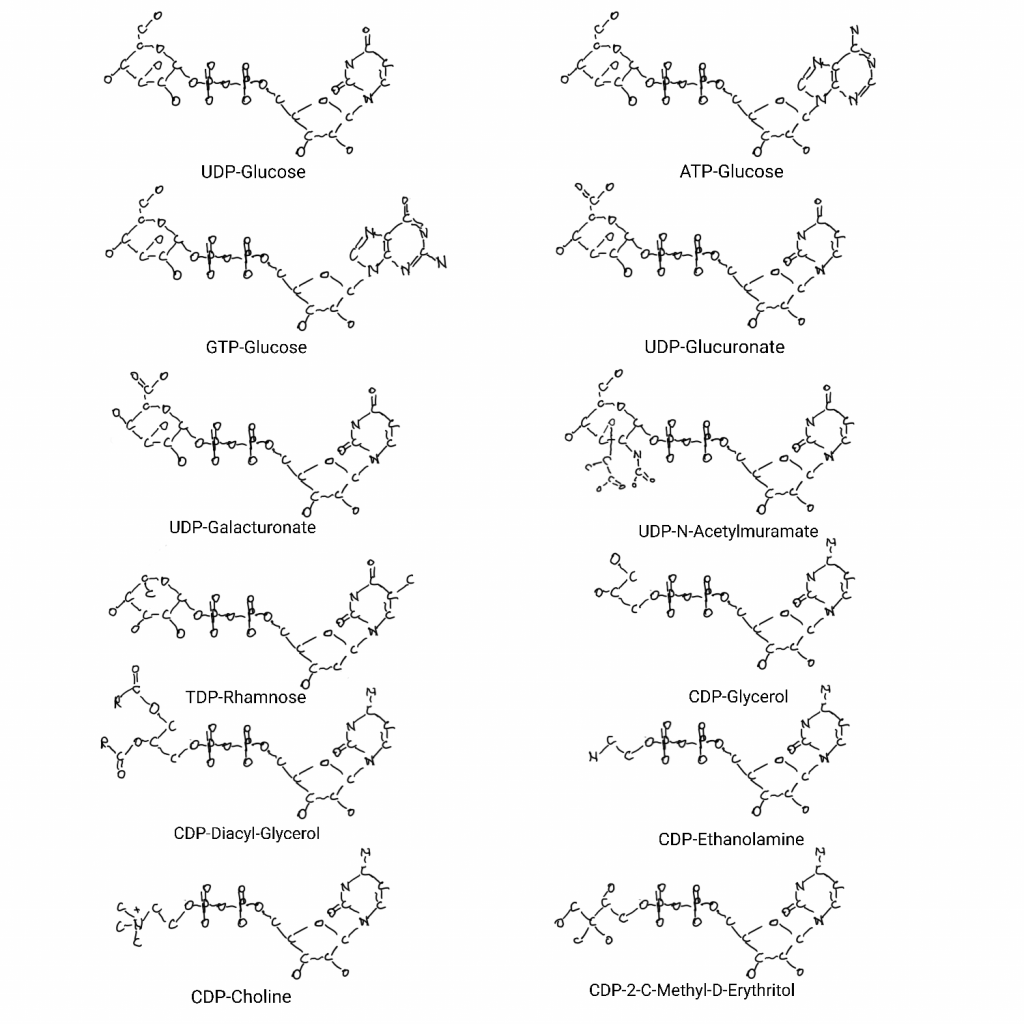
The 5th topology bin is S-Adenosyl-Methionine-dependent (SAM) methyltransferases. This also contains many families and the adenine-ribose-phosphate would be a common denominator for the first 5 bins so far. SAM is not just for methyl transfer though, the adenine-ribose can be an “adenylyl” modification (part of the molybdenum cofactor biosynthesis, sulfur carrier step). Most of methionine can be bound to something as homoserine leaving methyl-sulfur-ribose-adenine as well. As far as I can see this domain is all about SAM methyl transfer.
Tubulin nucleotide binding domain-like is very interesting. The cytoskeleton uses GTP hydrolysis on a timer to determine if the subunits like actin are monomers or polymers. And actin is an intracellular highway for cell contents.
The NagB/RpiA/CoA transferase-like domain has a lot of interesting things. Some transcription repressors, 5, 10-methlenyltetrahydrofolate synthetase. Related to fatty acid synthase there are coenzyme-A transferases and acetyl-CoA hydrolase (makes acetate). Citrate lyase is in the TCA cycle. IF-2B is a translation initiation factor. Finally there is glucosamine-6P isomerase and very interestingly ribose-5P isomerase. That interconverts ribose and ribulose in the pentose phosphate pathway.
The MurCD/PglD bin contains things that work with UDP-sugars and derivatives. MurC and MurD attach an alanine and a glycine to UDP-N-acetylmurmate in making bacterial cell walls.
The activating enzymes of ubiquitin-like proteins, or “E1 enzymes” are mostly studied in eukaryotes where ubiquitin targets proteins for degradation in the proteasome. But the general machinery exists in prokaryotes too and the activation process involves an adenylyl modification of the ubiquitin-like protein (UBL), where the adenylyl is replaced by the E1 (E1-UBL link) prior to attachment of the UBL to another protein, “activation” of the UBL. ThiF (thiamine biosynthesis) is in here, and it activates ThiS (on the big drawing) in a similar manner to exchange the adenylate with sulfur. That is something I want to add to the big drawing like I did with molybdenum cofactor biosynthesis, and MoeB (molybdenum cofactor biosynthesis) is in this bin too. No NAD(P)/FAD/SAM in here.
Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism Cappadocia 2017
The formate/glycerate dehydrogenase catalytic domain-like topology bin is interesting because every catalytic region could be relevant, and especially for smaller, more origins relevant molecules. There is S-adenosyl-l-homocysteine Hydrolase (methionine biosynthesis), and alanine dehydrogenase (alanine to pyruvate). Lysine oxioreductase, and dipicolinate synthase both in lysine metabolism. Glycerate, formate and lactate dehydrogenases. Also something called nicotinamide nucleotide Transhydrogenase. All of these require NAD(P) so it this may still just be cofactor binding regions.
The UDPG/MGDP dehydrogenase C-term topology bin doesn’t have much but it’s all nucleotide-sugar proteins. UDP-glucose is central to carbohydrates and polymers of them for storage and membrane parts. And at least one requires NAD.
Finally there are the aspartate/ornathine carbamoyl transferases. PyrB is in the pyrimidine pathway.
Stage 2.2: moving bicarbonate over 2 atoms.
PurE1 doesn’t use phosphate as mutase reactions change the structure of a molecule without adding or removing things, it’s a low energy process and doesn’t need ATP. The proteins and their various amino acid side chains can hold the molecule just right, and alter charge distributions such that the bicarbonate moves over. And this a protein domain family that I’ve already covered, the A+B3LS/Flavodoxin-like/GATase-like domain is the entire protein.
And bicarbonate is finally something in common with the pyrimidine pathway. Bicarbonate would have been in primordial oceans and would have seeped into the rock with water as well as contact with the outside of hydrothermal vent minerals.

Each level in geometry dash meltdown presents a unique set of obstacles and challenges, ensuring that the gameplay remains fresh and engaging throughout my entire journey across the various stages.