My students are also blogging here:
Today we talked about gap genes and a little bit about pair rule genes in flies, and to introduce the topic I summarized genetic epistasis. Epistasis is a fancy word for the interactions between genes, and we’ve already discussed it on the simplest level. You can imagine that a gene A, when expressed, activates the expression of gene B. The arrow in this diagram? That’s epistasis.
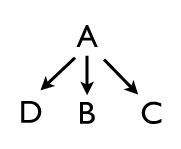
So far, so simple. This could describe how bicoid activates zygotic hunchback for instance. But of course not all epistatic interactions are linear and one dimensional; often one transcription factor will turn on or repress multiple genes — so A might switch on genes B, C, and D.
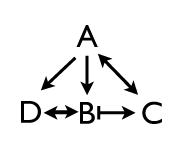
But wait! Now there is the potential for all kinds of combinatorial interactions: maybe C has positive feedback back on A, and B activates D and C, and D activates B, and C represses B. There’s a whole mathematically bewildering world of possibility here.
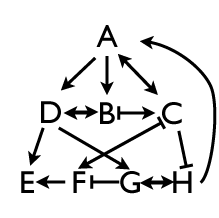
And it gets worse and worse. B, C, and D could have downstream effects on other genes, like E, F, G, and H, and each of those interact with each other and can have feedback effects as well. It’s not at all uncommon to be taking apart the sequence of events of a developmental pathway and discover a whole tangled snarl of epistatic interactions that lead to complicated patterns of gene expression.
And that’s molecular geneticists and developmental biologists do: they try to tease apart the snarl, asking how each gene interacts with all the other genes in the system, working out the kind of genetic circuitry shown in those diagrams. Often the approach is take it one gene at a time: knock out F, for instance, and ask what happens to the expression patterns of A, B, C, D, E, G, and H. Or upregulate D, and ask what all those other genes do. If you like logic puzzles, you’ll love epistatic studies, because that’s what they are: grand complicated logic puzzles with multiple cascading effects and usually only partial knowledge about what each component does. You’ll either have great fun with it all, or cultivate great headaches.
So most of the class hour was spent going through examples of these puzzles. The gap genes, for instance, are expressed in broad stripes in the embryo, and we can try to decipher the rules that establish the boundaries by taking out components. If hunchback is deleted, what do the giant, krüppel, and knirps stripes look like? Take out krüppel, what happens to knirps? So I led them through this series of experiments, asking them to come up with general rules regulating the expression of each stripe, and then using those rules to predict what would happen if we did a different experiment. I think they mostly got it.
But of course the discussion today was mostly about the gap genes, which are the second tier of genetic interactions (analogous to my third figure above). Next I introduced the pair rule genes, the third tier, rather like my fourth diagram. These are genes that are expressed in alternating stripes corresponding to parasegments in the fly…so we’ve gone from a few broad stripes to many narrow stripes. Each of those stripes, too, is independently regulated, with distinct control regions for each.
The real nightmare begins in the next class, when we start taking apart the many ways all of the pair rule genes interact with each other, and how their position is established partly by regulation by the gap genes and partly by mutual sorting out with combinations of activating and repressing interactions. It’s going to be loads of fun!

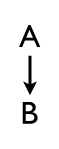

As I said before (Nah, PZ said) a lot of PEEK() and POKE() commands.
So these gap genes are the ones found in creationist fossils?
And here I thought that epistaxis was nosebleed.
Yuck, yuck
You may want to have a look at this paper (also Drosophila)
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1449574/
for a demonstration how these interactions are not static, but change with the genetic manipulation. Genetic networks can reconfigure to reveal a new topology after genes have been removed or altered in th network.
I keep reading this post as “…heavy on the epistaxis.” Which would be something different entirely.
Love this stuff, especially since it is a great excuse to set up big computational simulations. I know they do this at UWashington. When I was an undergrad bio student 35 years ago I had the temerity to suggest computational simulations of genetics and evolution to my profs. Essentially they patted me on the head and sent me back into the lab.
In the third diagram I see an arrow going from B to C. Yet in the description “C represses B”. To me it looks like a mistake. Or am I missing something?
B promotes C, while C represses B. There’s that –| going from C to B.
B–>C
C–|B
Put these together, and you get B|–>C.
I love reading these dev bio posts. Not that I dislike the rest of the blog, but I’m trained as a biochemist and worked in a molecular bio lab, so I really appreciate hearing things that aren’t my forte but within my area of interest. It’s like a little talk! I get so much from these, I love it. Thanks PZ.