Ammonia, glycine, and formate formed a ring on ribose-P, bicarbonate bound a nitrogen and was moved over 2 atoms. And little but phosphate was required for all these steps (externally, the protein still affects the process).
It’s worth mentioning here that this is another place where purine biosynthesis splits and joins another metabolic pathway. Previously thiamine and B12 joined at the first ring closure. Here aspartate is going to bind the nitrogen that used to be a glycine carbonyl, and then most of that aspartate will be removed as fumarate effectively making this a 2-step equivalent to glutamine as an ammonia dispenser. Aspartate as primordial ammonia dispenser?
After the removal of aspartate the product AICAR is a product of histidine biosynthesis as the big drawing shows, or should show if I remembered to draw an arrow (above). That explains why most of the molecule made with ATP and PRPP suddenly disappeared. Histidine is considered to be a late evolving amino acid, and AICAR feeds into purine biosynthesis instead of purine biosynthesis feeding into the connected metabolic pathway. Maybe it’s as simple as early purine biosynthesis being old and late purine biosynthesis being younger? Maybe one can just read age into the pathways somewhere.
Stage 3.1: addition of aspartate.
This is a protein domain family that I’ve already covered in general, but it has a few relations worth mentioning. SAICAR synthase, PurC, is related to the ATPgrasp domain, and a bunch of serine/threonine/tyrosine protein kinases. And phosphate is used in attaching the nitrogen of aspartate to the bicarbonate moved in the previous step by PurE1 (the phosphate attaches to CAIR by the bicarbonate first, and is replaced by aspartate).
While the ATPgrasp and protein kinases are topology siblings, PurC has some family siblings and they are all inositol kinases.
Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, Inositol-trisphosphate 3-kinase A, and Inositol-pentakisphosphate 2-kinase. This one may not be that origins related as inositol is not used by many prokaryotes. It’s often a membrane lipid modification, and the various forms of phosphorylated inositol can act as signalling molecules on and off the lipids. Since inositol can have 6 phosphates it has been used as phosphate storage in plants. I tend to think inositol isn’t something I should pursue with respect to origins but that could change.
Otherwise what is interesting is that with the arrival of aspartate we have all of the basic ingredients for pyrimidines, bicarbonate and aspartate (and ammonia and phosphate).
The reactions from PurK to PurC are a place to root thinking about the appearance of a second family of nucleotides, and the identity of any early purines with a single ring with multiple extensions as the pathway progresses. Did NCAIR and CAIR ever extend as nucleotides themselves? AIR? Was a SAICAR ever part of a chain of nucleotides?
And all that has been needed chemically for purines was a phosphate. And phosphate control evolved early. Earlier than purine biosynthesis directly. The following paper looked at kinds of metabolism and parts of purine metabolism relating to phosphate control are among the oldest parts.
Structural Phylogenomics Reveals Gradual Evolutionary Replacement of Abiotic Chemistries by Protein Enzymes in Purine Metabolism. Caetano-Anollés 2013
The following refers to Figure 1 in the results, the image is too large for WordPress to let me upload.
The most ancient enzymes (colored in deep red) are located in a horizontal transect of the subnetwork that is responsible for nucleotide interconversion, supporting previous evidence that metabolism originated in enzymes harboring the P-loop hydrolase fold and responsible for these kinase functions
Stage 3.2: aspartate leaves as fumarate, leaves its nitrogen.
No phosphate is needed for PurB to do its work. Stage 3 is a 2-step version of what the glutamine ammonia dispensers did previously. This way of delivering nitrogen occurs in arginine biosynthesis too. I can’t help but think of an aspartate accumulation step in history, between that and all of the other things on the big drawing that use or start with aspartate.
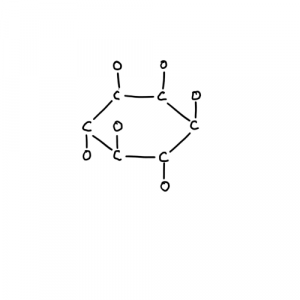
And what is fumarate? Fumerate is an intermediate in the TriCarboxylic Acid (TCA) cycle,or “citric acid cycle” or “Krebs cycle”.
The pyruvate and 2-oxogluterate dehydrogenase complexes are multi protein macromolecular complexes where carbon skeletons are passed from thiamine to lipoamide to coenzyme-A. I think part of our history involves being macromolecular complexes in mineral systems.
This cycle is where the precursor to glutamate comes from (2-oxogluterate), and the precursor to aspartate is involved (oxaloacetate). Add glycine at succinyl-CoA and you get the pathway leading to heme and chlorophyll (not shown). If I had to stereotype the TCA cycle it would be about converting a 4C (oxaloacetate) and a 2C (acetyl-CoA) into a 5C (2-oxogluterate). It also produces protons via NAD(P) (can be used to regenerate ATP), and GTP/ATP from phosphate and GDP/ADP. So the removal of fumerate may tie purines to the TCA cycle. This cycle has a reverse version like glycolysis and gluconeogenesis, there are separate enzymes that do irreversible steps (arrows that just go in one direction).
Fumerate also works as an electron acceptor in electron transport chains.
As for the protein domains making up PurB, the situation is reminiscent of PurM. PurB is divided into 3 subdomains and most of the same proteins are in all 3. But the 3 subdomains may be more closely related to each other with PurB, they are all in the same architecture bin where the N and C ends of PurM are in different architecture bins.
(As I type this ECOD seems to have a problem where it somehow has Alpha Arrays architecture in the Beta Barrels bin. When I first did this all 3 domains were in Alpha Arrays, now the C-terminal region is in Beta Barrels. There are no barrels made from Beta Sheets in this domain so I will keep it as Alpha Arrays (arrays of Alpha Helices). There are a few mistakes on the big drawing due to this issue which I will correct. Specifically some Beta Barrels are actually Alpha Arrays. Otherwise the relatives of this domain all seem there.)
PurB is in a group called “L-Aspartase-like” and there are “L-Aspartase-like-N”, “L-Aspartase-like-M” (middle), and “L-Aspartase-like-C” X bins. In these bins are aspartase (useful under nitrogen starvation). Histidine ammonia lyase (histidine breakdown), phenylalanine ammonia lyase (plants) tyrosine and phenylalanine amino mutases (makes amino acids of opposite chirality) Adenylosuccinate lyase (PurB), delta crystallin (eye proteins), and fumerate hydratase (TCA cycle protein). The exceptions seem to be 3-carboxy-cis,cis-muconate cycloisomerase (benzoate degradation) absent from the N-terminal end, and the histidine, phenylalanine, and tyrosine proteins seem absent at the C-terminal end but I need to look at things with another archive to be sure.
Stage 4.1: formylation of the aspartate derived nitrogen.
This stage and the following step is a place where life has more than one path. In some organisms the bifunctional PurHJ, (sometimes just called PurH) carries out the steps sequentially. In other organisms PurP and PurO carry out the steps. The following paper covers how widespread these proteins are distributed among prokaryotes and the authors named the PurH associated formylation protein PurV and the following protein PurJ when the functions are in separate proteins.
PurJ is in the A+B3LS architecture bin, and the MGS-like X, homology, and topology bins (methylglyoxal synthase) so these are all related. PurJ is listed in ECOD as bifunctional protein PurH. CarB of pyrimidine biosynthesis is in here too. Lastly there is methylglyoxal synthase which provides an alternate way to metabolize 3C-phosphates when phosphate is limiting. Notably PurV activity does not require phosphate but does use folate like PurN.
PurP is familiar because it is an ATPgrasp enzyme. It also has the preATPgrasp domain, but not the C-terminal CODH-MO-N-like domain that I find so interesting. So similarly to Stage 1 life has ways of formylating with and without phosphate. But in this case PurP seems to be limited to archaea. The above paper found no bacteria using PurP. Most bacteria use PurH (the whole thing) and some have separate PurJ and PurV/PurO. Some bacteria and archaea have no identified proteins for Stage 4, autocatalytic?
Stage 4.2: closure of the second ring and the formation of IMP.
PurV is in the A+B3L architecture bin (not a sandwich), and cytidine deaminase-like X, homology and topology bins. There are a lot of proteins in here and modification of poly RNA or DNA seems to be a theme, editing enzymes. There is a whole level where RNA can be edited after transcription to change protein sequence or make other function changes. I’m amused by the tRNA adenosine deaminase TadA (TA-DA!). There are other adenine deaminases. There are cytosine deaminases, and guanosine deaminases. There is a pre-mRNA splicing factor, Prp8. RadC which is thought to be a nuclease (cuts polynucleotides).
Isopeptidase activity is associated with this group. An isopeptidase is a protein that cuts a protein outside of the main protein sequence, the relevant example here being ubiquitin-like proteins that are attached to a lysine residue (or attached to another side chain in other isopeptidases). This system is where ubiquitin tags proteins destined to be broken down in the proteasome (the cellular trash can) is eukaryotic but is related to systems in bacteria and archaea. Prokaryotic relatives of the ubiquitin system is something I will need to read more about.
LpxL is in here and that is a UDP-2,3-diacylglucosamine pyrophosphatse (removes diphosphate at some point). That is interesting due to sugars and lipid related things bound to nucleotide handles in the previous post, but is a minor bit of chemistry in this group.
FdhD is interesting. That is a sulfur carrier required for the function of formate dehydrogenase. This isn’t the first sulfur carrier/ubiquitin-like system connection I have seen. ThiS and MoaD are ubiquitin-like. Maybe there is a sulfur carrier link to isopeptidase-like (deubiquitinase-like) proteins as well?
PurO
PurO is in a group I have already covered, the A+B4L architecture bin, the NTN/PP2C X bin, and NTN (N-Terminal amiNohydrolase) homology bin. It’s a bit like coming full circle at Inosine MonoPhosphate (IMP) since PurF was at the beginning and contains one of these domains. PurO is in the proteasome subunits family bin. This is interesting because while PurO joins parts to make a ring, proteasome subunits cut proteins.
Now with second ring closure something that gets an abbreviation like AGCT, I, IMP. I is never found in transcription directly, but some of those editing enzymes can turn something into I. That affects codon base pairing in translation leading to different effects. tRNAs can contain I.
Nucleotides as intracellular signalling molecules.
Inosine is also an example of a whole level I haven’t read about and integrated into my thinking yet, it is an intracellular signalling molecule like cyclic AMP or GMP derivatives. AICAR is a signalling molecule too. Guanosine tetraphosphate is a signalling molecule.
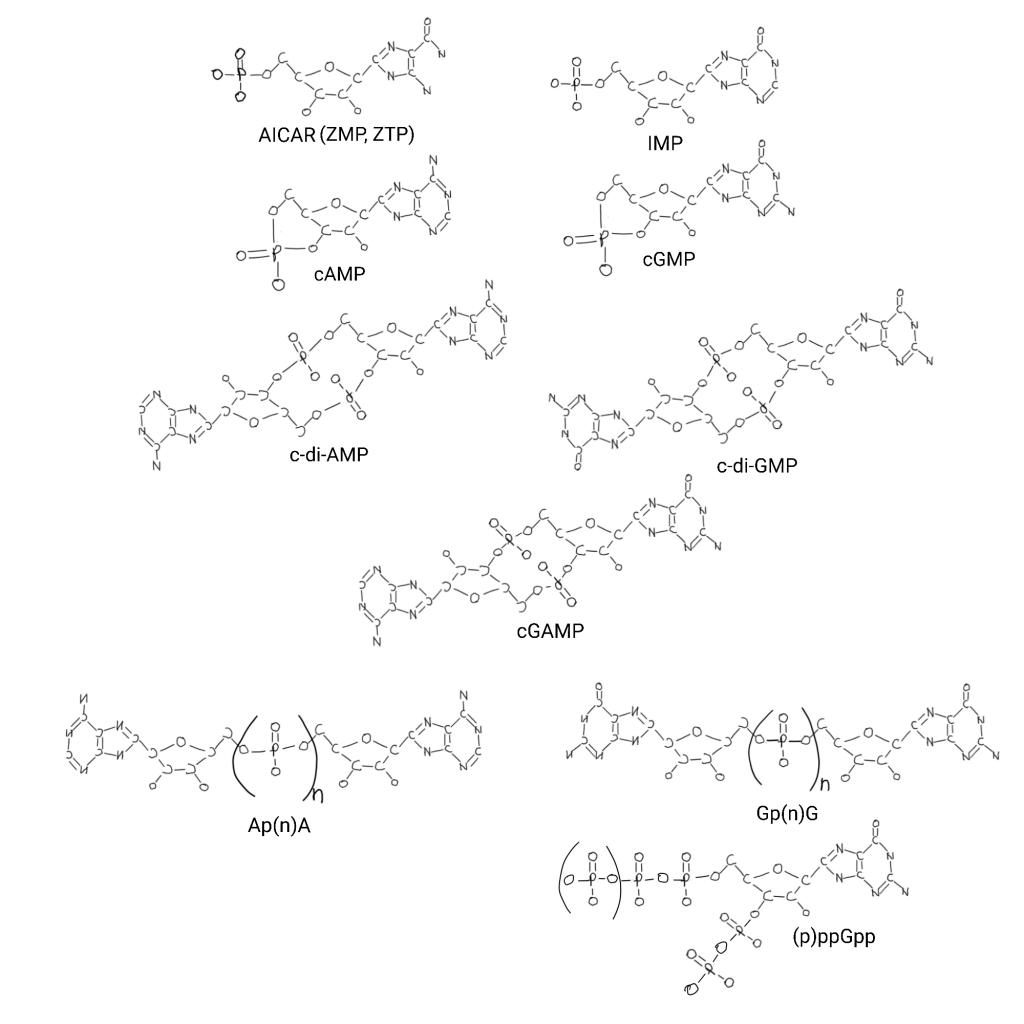
And these are all signalling molecules (“alarmone” is used with many of these) in addition to AICAR (called ZMP and ZTP as signalling molecules) and IMP. ZTP and IMP act in regulation of steps of purine biosynthesis. I found a paper looking for these systems in some archaea with a big table.
Many are thought to be ancient.
Alarmones as Vestiges of a Bygone RNA World. Hernández-Morales 2019
Cyclic AMP (cAMP) shifts metabolism in response to things. In bacteria it is high when things other than glucose are carbon sources. Glucose is like an ancestral default sugar maybe. cAMP has many other functions like osmotic pressure and DNA damage. The rest of them are hard to find stereotyped roles for. It’s the cutting edge of this research.
Cyclic GMP (cGMP) is involved in UV stress adaptation and cyst (suspended animation) formation. There is a cIMP, cCMP and cUMP too but very little is known about their role.
c-di-AMP is also involved in osmotic pressure. c-di-GMP is involved in transition from motile to sessile living. cGAMP is only found in metazoa, so no origins role. But I drew a 2,3 cGAMP and there is a 3,3 cGAMP that is involved in viral infection.
Ap(n)A has different forms and Ap4A is best known. Ap4A is an alarmone for many stress conditions. I couldn’t find anything on Gp(n)G. (p)ppGpp is also involved in stress signalling.
That’s it for stages 3, 4 and an area of nucleotide biochemistry I still need to look at.

The level of detail and the connections you draw between purine biosynthesis, protein domains, and evolutionary pathways really show strong expertise. It’s challenging but rewarding to read, especially for readers interested in biochemical evolution.
I also enjoy how Dinosaur Game changes visuals with day and night mode. It’s a small detail, but it keeps the experience from feeling boring during longer runs.
what these cyclic nucleotides do?